Uncovering pathways in DNA oligonucleotide hybridization via transition state analysis
- PMID: 19815517
- PMCID: PMC2759370
- DOI: 10.1073/pnas.0904721106
Uncovering pathways in DNA oligonucleotide hybridization via transition state analysis
Erratum in
- Proc Natl Acad Sci U S A. 2009 Dec 8;106(49):21007
Abstract
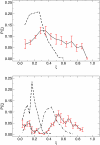
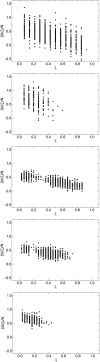
DNA hybridization plays a central role in biology and, increasingly, in materials science. Yet, there is no precedent for examining the pathways by which specific single-stranded DNA sequences interact to assemble into a double helix. A detailed model of DNA is adopted in this work to examine such pathways and to determine the role of sequence, if any, on DNA hybridization. Transition path sampling simulations reveal that DNA rehybridization is prompted by a distinct nucleation event involving molecular sites with approximately four bases pairing with partners slightly offset from those involved in ideal duplexation. Nucleation is promoted in regions with repetitive base pair sequence motifs, which yield multiple possibilities for finding complementary base partners. Repetitive sequences follow a nonspecific pathway to renaturation consistent with a molecular "slithering" mechanism, whereas random sequences favor a restrictive pathway involving the formation of key base pairs before renaturation fully ensues.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Sequence effects in the melting and renaturation of short DNA oligonucleotides: structure and mechanistic pathways.J Phys Condens Matter. 2009 Jan 21;21(3):034105. doi: 10.1088/0953-8984/21/3/034105. Epub 2008 Dec 17. J Phys Condens Matter. 2009. PMID: 21817250 Free PMC article.
-
Kinetics and dynamics of DNA hybridization.Acc Chem Res. 2011 Nov 15;44(11):1172-81. doi: 10.1021/ar200068j. Epub 2011 Jun 30. Acc Chem Res. 2011. PMID: 21718008
-
Sequence-Dependent Mechanism of DNA Oligonucleotide Dehybridization Resolved through Infrared Spectroscopy.J Am Chem Soc. 2016 Sep 14;138(36):11792-801. doi: 10.1021/jacs.6b05854. Epub 2016 Aug 31. J Am Chem Soc. 2016. PMID: 27519555
-
Oligonucleotides with Cationic Backbone and Their Hybridization with DNA: Interplay of Base Pairing and Electrostatic Attraction.Chemistry. 2018 Feb 1;24(7):1544-1553. doi: 10.1002/chem.201704338. Epub 2017 Dec 27. Chemistry. 2018. PMID: 29048135 Free PMC article.
-
Mismatch formation in solution and on DNA microarrays: how modified nucleosides can overcome shortcomings of imperfect hybridization caused by oligonucleotide composition and base pairing.Mol Biosyst. 2008 Mar;4(3):232-45. doi: 10.1039/b713259j. Epub 2008 Feb 4. Mol Biosyst. 2008. PMID: 18437266 Review.
Cited by
-
The power of coarse graining in biomolecular simulations.Wiley Interdiscip Rev Comput Mol Sci. 2014 May;4(3):225-248. doi: 10.1002/wcms.1169. Wiley Interdiscip Rev Comput Mol Sci. 2014. PMID: 25309628 Free PMC article.
-
DNA hybridization kinetics: zippering, internal displacement and sequence dependence.Nucleic Acids Res. 2013 Oct;41(19):8886-95. doi: 10.1093/nar/gkt687. Epub 2013 Aug 8. Nucleic Acids Res. 2013. PMID: 23935069 Free PMC article.
-
Presentation of large DNA molecules for analysis as nanoconfined dumbbells.Macromolecules. 2013 Oct 22;46(20):8356-8368. doi: 10.1021/ma400926h. Macromolecules. 2013. PMID: 24683272 Free PMC article.
-
A coarse-grained model of DNA with explicit solvation by water and ions.J Phys Chem B. 2011 Jan 13;115(1):132-42. doi: 10.1021/jp107028n. Epub 2010 Dec 14. J Phys Chem B. 2011. PMID: 21155552 Free PMC article.
-
Determining Sequence-Dependent DNA Oligonucleotide Hybridization and Dehybridization Mechanisms Using Coarse-Grained Molecular Simulation, Markov State Models, and Infrared Spectroscopy.J Am Chem Soc. 2021 Oct 27;143(42):17395-17411. doi: 10.1021/jacs.1c05219. Epub 2021 Oct 13. J Am Chem Soc. 2021. PMID: 34644072 Free PMC article.
References
-
- Voet D, Voet JG, Pratt CW. Fundamentals of Biochemistry. New York: Wiley; 1999. pp. 751–930.
-
- Saiki RK, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. - PubMed
-
- Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;404:297–302. - PubMed
-
- Freifelder D. Principles of Physical Chemistry with Applications to the Biological Sciences. Boston: Jones and Bartlett; 1985. pp. 513–517.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

