Equine rhinitis A virus and its low pH empty particle: clues towards an aphthovirus entry mechanism?
- PMID: 19816570
- PMCID: PMC2752993
- DOI: 10.1371/journal.ppat.1000620
Equine rhinitis A virus and its low pH empty particle: clues towards an aphthovirus entry mechanism?
Abstract
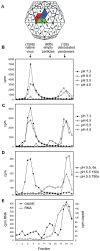
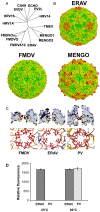
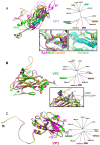
Equine rhinitis A virus (ERAV) is closely related to foot-and-mouth disease virus (FMDV), belonging to the genus Aphthovirus of the Picornaviridae. How picornaviruses introduce their RNA genome into the cytoplasm of the host cell to initiate replication is unclear since they have no lipid envelope to facilitate fusion with cellular membranes. It has been thought that the dissociation of the FMDV particle into pentameric subunits at acidic pH is the mechanism for genome release during cell entry, but this raises the problem of how transfer across the endosome membrane of the genome might be facilitated. In contrast, most other picornaviruses form 'altered' particle intermediates (not reported for aphthoviruses) thought to induce membrane pores through which the genome can be transferred. Here we show that ERAV, like FMDV, dissociates into pentamers at mildly acidic pH but demonstrate that dissociation is preceded by the transient formation of empty 80S particles which have released their genome and may represent novel biologically relevant intermediates in the aphthovirus cell entry process. The crystal structures of the native ERAV virus and a low pH form have been determined via highly efficient crystallization and data collection strategies, required due to low virus yields. ERAV is closely similar to FMDV for VP2, VP3 and part of VP4 but VP1 diverges, to give a particle with a pitted surface, as seen in cardioviruses. The low pH particle has internal structure consistent with it representing a pre-dissociation cell entry intermediate. These results suggest a unified mechanism of picornavirus cell entry.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Equine Rhinitis A Virus Mutants with Altered Acid Resistance Unveil a Key Role of VP3 and Intrasubunit Interactions in the Control of the pH Stability of the Aphthovirus Capsid.J Virol. 2016 Oct 14;90(21):9725-9732. doi: 10.1128/JVI.01043-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27535044 Free PMC article.
-
Cell entry of the aphthovirus equine rhinitis A virus is dependent on endosome acidification.J Virol. 2010 Jun;84(12):6235-40. doi: 10.1128/JVI.02375-09. Epub 2010 Apr 7. J Virol. 2010. PMID: 20375159 Free PMC article.
-
Internal ribosomal entry site-mediated translation initiation in equine rhinitis A virus: similarities to and differences from that of foot-and-mouth disease virus.J Virol. 2000 Dec;74(24):11708-16. doi: 10.1128/jvi.74.24.11708-11716.2000. J Virol. 2000. PMID: 11090170 Free PMC article.
-
Equine picornaviruses: well known but poorly understood.Vet Microbiol. 2013 Nov 29;167(1-2):78-85. doi: 10.1016/j.vetmic.2013.05.012. Epub 2013 Jun 10. Vet Microbiol. 2013. PMID: 23820049 Review.
-
Foot and mouth disease.Res Vet Sci. 2002 Dec;73(3):195-9. doi: 10.1016/s0034-5288(02)00105-4. Res Vet Sci. 2002. PMID: 12443674 Review.
Cited by
-
Molecular basis for the different PCV2 susceptibility of T-lymphoblasts in Landrace and Piétrain pigs.Vet Res. 2024 Feb 19;55(1):22. doi: 10.1186/s13567-024-01275-0. Vet Res. 2024. PMID: 38374131 Free PMC article.
-
The In Silico Prediction of Hotspot Residues that Contribute to the Structural Stability of Subunit Interfaces of a Picornavirus Capsid.Viruses. 2020 Mar 31;12(4):387. doi: 10.3390/v12040387. Viruses. 2020. PMID: 32244486 Free PMC article.
-
The pH Stability of Foot-and-Mouth Disease Virus Particles Is Modulated by Residues Located at the Pentameric Interface and in the N Terminus of VP1.J Virol. 2015 May;89(10):5633-42. doi: 10.1128/JVI.03358-14. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762735 Free PMC article.
-
Structural Dynamics of Nonenveloped Virus Disassembly Intermediates.J Virol. 2019 Oct 29;93(22):e01115-19. doi: 10.1128/JVI.01115-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31484752 Free PMC article.
-
Capsid protein VP4 of human rhinovirus induces membrane permeability by the formation of a size-selective multimeric pore.PLoS Pathog. 2014 Aug 7;10(8):e1004294. doi: 10.1371/journal.ppat.1004294. eCollection 2014 Aug. PLoS Pathog. 2014. PMID: 25102288 Free PMC article.
References
-
- Newman JF, Rowlands DJ, Brown F. A physicochemical sub-grouping of the mammalian picornaviruses. J Gen Virol. 1973;18:171–180. - PubMed
-
- Newman JFE, Rowlands DJ, Brown F, Goodridge D, Burrows R, et al. Physicochemical characterization of two serologically unrelated rhinoviruses. Intervirology. 1977;8:145–154. - PubMed
-
- Studdert MJ, Gleeson LJ. Isolation and characterisation of an equine rhinovirus. Zentbl Vetmed B. 1978;25:225–237. - PubMed
-
- Wutz G, Auer H, Nowotny N, Grosse B, Skern T, et al. Equine rhinovirus serotypes 1 and 2: relationship to each other and to aphthoviruses and cardioviruses. J Gen Virol. 1996;77:1719–1730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

