Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals
- PMID: 19816605
- PMCID: PMC2756586
- DOI: 10.1371/journal.pone.0007370
Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals
Abstract
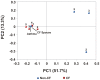
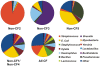
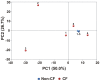
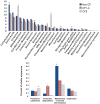
The human respiratory tract is constantly exposed to a wide variety of viruses, microbes and inorganic particulates from environmental air, water and food. Physical characteristics of inhaled particles and airway mucosal immunity determine which viruses and microbes will persist in the airways. Here we present the first metagenomic study of DNA viral communities in the airways of diseased and non-diseased individuals. We obtained sequences from sputum DNA viral communities in 5 individuals with cystic fibrosis (CF) and 5 individuals without the disease. Overall, diversity of viruses in the airways was low, with an average richness of 175 distinct viral genotypes. The majority of viral diversity was uncharacterized. CF phage communities were highly similar to each other, whereas Non-CF individuals had more distinct phage communities, which may reflect organisms in inhaled air. CF eukaryotic viral communities were dominated by a few viruses, including human herpesviruses and retroviruses. Functional metagenomics showed that all Non-CF viromes were similar, and that CF viromes were enriched in aromatic amino acid metabolism. The CF metagenomes occupied two different metabolic states, probably reflecting different disease states. There was one outlying CF virome which was characterized by an over-representation of Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase, an enzyme involved in the bacterial stringent response. Unique environments like the CF airway can drive functional adaptations, leading to shifts in metabolic profiles. These results have important clinical implications for CF, indicating that therapeutic measures may be more effective if used to change the respiratory environment, as opposed to shifting the taxonomic composition of resident microbiota.
Conflict of interest statement
Figures



References
-
- Heyder J. Deposition of Inhaled Particles in the Human Respiratory Tract and Consequences for Regional Targeting in Respiratory Drug Delivery. Proc Am Thorac Soc. 2004;1:315–320. - PubMed
-
- Corne JM, et al. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359:831–834. - PubMed
-
- Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology (Reading, Engl.) 2007;153:917–923. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

