Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs
- PMID: 19818023
- PMCID: PMC2917040
- DOI: 10.1111/j.1365-2958.2009.06895.x
Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs
Abstract
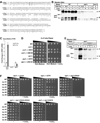
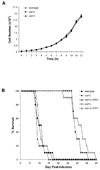
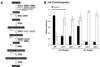
In the human fungal pathogen Cryptococcus neoformans, the SREBP orthologue Sre1 is important for adaptation and growth in nutrient-limiting host tissues. In this study, we characterize the C. neoformans serotype A Sre1 and its activating protease, Stp1. We demonstrate that Stp1 is a functionally conserved orthologue of the mammalian Site-2 protease and that Stp1 cleaves Sre1 within its predicted first transmembrane segment. Gene expression analysis revealed that Stp1 is required for both Sre1-dependent and Sre1-independent gene transcription, indicating that other substrates of Stp1 may exist. Using gas chromatography, we showed that Sre1 and Stp1 are required for both normoxic and hypoxic ergosterol biosynthesis, and therefore cells lacking SRE1 or STP1 are defective for growth in the presence of low levels of the ergosterol biosynthesis inhibitors, itraconazole and 25-thialanosterol. Importantly, our studies demonstrated fungicidal effects of itraconazole and 25-thialanosterol towards sre1Delta and stp1Delta cells, demonstrating that the Sre1 pathway is required for both growth and survival in the presence of sterol biosynthesis-inhibiting antifungal drugs. Given the need for fungicidal drugs, we propose that inhibitors of Stp1, Sre1, or other regulators of Sre1 function administered in combination with a sterol synthesis inhibitor could prove an effective anticryptococcal therapy.
Figures



References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. - PMC - PubMed
-
- Bennett JE, Kwon-Chung KJ, Howard DH. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am.J.Epidemiol. 1977;105:582–586. - PubMed
-
- Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol.Microbiol. 2007;64:614–629. - PubMed
-
- Chow ED, Liu OW, O'Brien S, Madhani HD. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: nitric oxide stress and body temperature. Current Genetics. 2007;52:137–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

