Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases
- PMID: 19818708
- PMCID: PMC2847577
- DOI: 10.1016/j.molcel.2009.09.022
Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases
Abstract
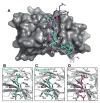
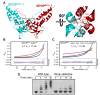
In the largest E3 ligase subfamily, Cul3 binds a BTB domain, and an associated protein-interaction domain such as MATH recruits substrates for ubiquitination. Here, we present biochemical and structural analyses of the MATH-BTB protein, SPOP. We define a SPOP-binding consensus (SBC) and determine structures revealing recognition of SBCs from the phosphatase Puc, the transcriptional regulator Ci, and the chromatin component MacroH2A. We identify a dimeric SPOP-Cul3 assembly involving a conserved helical structure C-terminal of BTB domains, which we call "3-box" due to its facilitating Cul3 binding and its resemblance to F-/SOCS-boxes in other cullin-based E3s. Structural flexibility between the substrate-binding MATH and Cul3-binding BTB/3-box domains potentially allows a SPOP dimer to engage multiple SBCs found within a single substrate, such as Puc. These studies provide a molecular understanding of how MATH-BTB proteins recruit substrates to Cul3 and how their dimerization and conformational variability may facilitate avid interactions with diverse substrates.
Figures


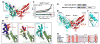


References
-
- Allen E, Ding J, Wang W, Pramanik S, Chou J, Yau V, Yang Y. Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature. 2005;438:224–228. - PubMed
-
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006a;443:590–593. - PubMed
-
- Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, Moon RT. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006b;8:348–357. - PubMed
-
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
-
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- GM070565/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- R01GM069530/GM/NIGMS NIH HHS/United States
- CA92584/CA/NCI NIH HHS/United States
- P41 RR015301/RR/NCRR NIH HHS/United States
- P30CA021765/CA/NCI NIH HHS/United States
- RR-15301/RR/NCRR NIH HHS/United States
- R01 GM069530/GM/NIGMS NIH HHS/United States
- P01 CA092584/CA/NCI NIH HHS/United States
- R01 GM070565/GM/NIGMS NIH HHS/United States
- DRG 2021-9/HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

