SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation
- PMID: 19818711
- PMCID: PMC2766281
- DOI: 10.1016/j.molcel.2009.08.015
SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation
Abstract
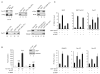
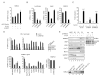
The Ski-interacting protein SKIP/SNW1 associates with the P-TEFb/CDK9 elongation factor and coactivates inducible genes, including HIV-1. We show here that SKIP also associates with c-Myc and Menin, a subunit of the MLL1 histone methyltransferase (H3K4me3) complex and that HIV-1 Tat transactivation requires c-Myc and Menin, but not MLL1 or H3K4me3. RNAi-ChIP experiments reveal that SKIP acts downstream of Tat:P-TEFb to recruit c-Myc and its partner TRRAP, a scaffold for histone acetyltransferases, to the HIV-1 promoter. By contrast, SKIP is recruited by the RNF20 H2B ubiquitin ligase to the basal HIV-1 promoter in a step that is bypassed by Tat and downregulated by c-Myc. Of interest, we find that SKIP and P-TEFb are dispensable for UV stress-induced HIV-1 transcription, which is strongly upregulated by treating cells with the CDK9 inhibitor flavopiridol. Thus, SKIP acts with c-Myc and Menin to promote HIV-1 Tat:P-TEFb transcription at an elongation step that is bypassed under stress.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

