Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks
- PMID: 19819872
- PMCID: PMC2797203
- DOI: 10.1074/jbc.M109.065730
Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks
Abstract
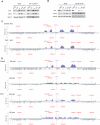
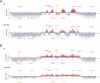
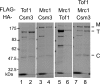
Mrc1 (mediator of replication checkpoint), Tof1 (topoisomerase I interacting factor), and Csm3 (chromosome segregation in meiosis) are checkpoint-mediator proteins that function during DNA replication and activate the effector kinase Rad53. We reported previously that Mrc1 and Tof1 are constituents of the replication machinery and that both proteins are required for the proper arrest and stabilization of replication forks in the presence of hydroxyurea. In our current study, we show that Csm3 is a component of moving replication forks and that both Tof1 and Csm3 are specifically required for the association of Mrc1 with these structures. In contrast, the deletion of mrc1 did not affect the association of Tof1 and Csm3 with the replication fork complex. In agreement with previous observations in yeast cells, the results of a baculovirus coexpression system showed that these three proteins interact directly with each other to form a mediator complex in the absence of replication forks.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

