Identification of defense compounds in Barbarea vulgaris against the herbivore Phyllotreta nemorum by an ecometabolomic approach
- PMID: 19819983
- PMCID: PMC2785962
- DOI: 10.1104/pp.109.136952
Identification of defense compounds in Barbarea vulgaris against the herbivore Phyllotreta nemorum by an ecometabolomic approach
Abstract
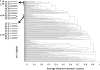
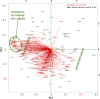
Winter cress (Barbarea vulgaris) is resistant to a range of insect species. Some B. vulgaris genotypes are resistant, whereas others are susceptible, to herbivory by flea beetle larvae (Phyllotreta nemorum). Metabolites involved in resistance to herbivory by flea beetles were identified using an ecometabolomic approach. An F2 population representing the whole range from full susceptibility to full resistance to flea beetle larvae was generated by a cross between a susceptible and a resistant B. vulgaris plant. This F2 offspring was evaluated with a bioassay measuring the ability of susceptible flea beetle larvae to survive on each plant. Metabolites that correlated negatively with larvae survival were identified through correlation, cluster, and principal component analyses. Two main clusters of metabolites that correlate negatively with larvae survival were identified. Principal component analysis grouped resistant and susceptible plants as well as correlated metabolites. Known saponins, such as hederagenin cellobioside and oleanolic acid cellobioside, as well as two other saponins correlated significantly with plant resistance. This study shows the potential of metabolomics to identify bioactive compounds involved in plant defense.
Figures



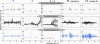



References
-
- Agerbirk N, Olsen CE, Bibby BM, Frandsen HO, Brown LD, Nielsen JK, Renwick JAA (2003. a) A saponin correlated with variable resistance of Barbarea vulgaris to diamondback moth Plutella xylostella. J Chem Ecol 29 1417–1433 - PubMed
-
- Agerbirk N, Olsen CE, Nielsen JK (2001) Seasonal variation in leaf glucosinolates and insect resistance in two types of Barbarea vulgaris ssp. arcuata. Phytochemistry 58 91–100 - PubMed
-
- Agerbirk N, Ørgaard M, Nielsen JK (2003. b) Glucosinolates, flea beetle resistance, and leaf pubescence as taxonomic characters in the genus Barbarea (Brassicaceae). Phytochemistry 63 69–80 - PubMed
-
- Bailey CD, Koch MA, Mayer M, Mummenhoff K, O'Kane SL Jr, Warwick SI, Windham MD, Al-Shehbaz IA (2006) Toward a global phylogeny of the Brassicaceae. Mol Biol Evol 23 2142–2160 - PubMed
-
- Blow N (2008) Metabolomics: biochemistry's new look. Nature 455 697–700 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

