Growth phase and (p)ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli
- PMID: 19820090
- PMCID: PMC2786607
- DOI: 10.1128/JB.00412-09
Growth phase and (p)ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli
Abstract
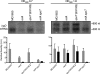
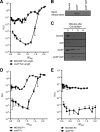
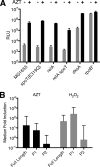
The antiadaptor protein IraD inhibits the proteolysis of the alternative sigma factor, RpoS, which promotes the synthesis of >100 genes during the general stress response and during stationary phase. Our previous results showed that IraD determines RpoS steady-state levels during exponential growth and mediates its stabilization after DNA damage. In this study, we show by promoter fusions that iraD was upregulated during the transition from exponential growth to stationary phase. The levels of RpoS likewise rose during this transition in a partially IraD-dependent manner. The expression of iraD was under the control of ppGpp. The expression of iraD required RelA and SpoT (p)ppGpp synthetase activities and was dramatically induced by a "stringent" allele of RNA polymerase, culminating in elevated levels of RpoS. Surprisingly, DksA, normally required for transcriptional effects of the stringent response, repressed iraD expression, suggesting that DksA can exert regulatory effects independent of and opposing those of (p)ppGpp. Northern blot analysis and 5' rapid amplification of cDNA ends revealed two transcripts for iraD in wild-type strains; the smaller was regulated positively by RelA during growth; the larger transcript was induced specifically upon transition to stationary phase and was RelA SpoT dependent. A reporter fusion to the distal promoter indicated that it accounts for growth-phase regulation and DNA damage inducibility. DNA damage inducibility occurred in strains unable to synthesize (p)ppGpp, indicating an additional mode of regulation. Our results suggest that the induction of RpoS during transition to stationary phase and by (p)ppGpp occurs at least partially through IraD.
Figures



References
-
- Aberg, A., V. Shingler, and C. Balsalobre. 2008. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 67:1223-1241. - PubMed
-
- Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

