DNaseI hypersensitivity at gene-poor, FSH dystrophy-linked 4q35.2
- PMID: 19820107
- PMCID: PMC2794184
- DOI: 10.1093/nar/gkp833
DNaseI hypersensitivity at gene-poor, FSH dystrophy-linked 4q35.2
Abstract
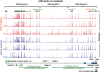
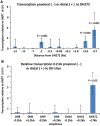
A subtelomeric region, 4q35.2, is implicated in facioscapulohumeral muscular dystrophy (FSHD), a dominant disease thought to involve local pathogenic changes in chromatin. FSHD patients have too few copies of a tandem 3.3-kb repeat (D4Z4) at 4q35.2. No phenotype is associated with having few copies of an almost identical repeat at 10q26.3. Standard expression analyses have not given definitive answers as to the genes involved. To investigate the pathogenic effects of short D4Z4 arrays on gene expression in the very gene-poor 4q35.2 and to find chromatin landmarks there for transcription control, unannotated genes and chromatin structure, we mapped DNaseI-hypersensitive (DH) sites in FSHD and control myoblasts. Using custom tiling arrays (DNase-chip), we found unexpectedly many DH sites in the two large gene deserts in this 4-Mb region. One site was seen preferentially in FSHD myoblasts. Several others were mapped >0.7 Mb from genes known to be active in the muscle lineage and were also observed in cultured fibroblasts, but not in lymphoid, myeloid or hepatic cells. Their selective occurrence in cells derived from mesoderm suggests functionality. Our findings indicate that the gene desert regions of 4q35.2 may have functional significance, possibly also to FSHD, despite their paucity of known genes.
Figures




References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

