Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy
- PMID: 19820108
- PMCID: PMC2794176
- DOI: 10.1093/nar/gkp816
Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy
Abstract
Important regions of rRNA are rich in nucleotide modifications that can have strong effects on ribosome biogenesis and translation efficiency. Here, we examine the influence of pseudouridylation and 2'-O-methylation on translation accuracy in yeast, by deleting the corresponding guide snoRNAs. The regions analyzed were: the decoding centre (eight modifications), and two intersubunit bridge domains-the A-site finger and Helix 69 (six and five modifications). Results show that a number of modifications influence accuracy with effects ranging from 0.3- to 2.4-fold of wild-type activity. Blocking subsets of modifications, especially from the decoding region, impairs stop codon termination and reading frame maintenance. Unexpectedly, several Helix 69 mutants possess ribosomes with increased fidelity. Consistent with strong positional and synergistic effects is the finding that single deletions can have a more pronounced phenotype than multiple deficiencies in the same region. Altogether, the results demonstrate that rRNA modifications have significant roles in translation accuracy.
Figures
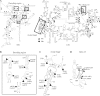

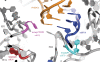

Similar articles
-
rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity.Mol Cell. 2007 Dec 28;28(6):965-77. doi: 10.1016/j.molcel.2007.10.012. Mol Cell. 2007. PMID: 18158895
-
Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing.RNA. 2009 Sep;15(9):1716-28. doi: 10.1261/rna.1724409. Epub 2009 Jul 23. RNA. 2009. PMID: 19628622 Free PMC article.
-
The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9.Nucleic Acids Res. 2016 Oct 14;44(18):8951-8961. doi: 10.1093/nar/gkw564. Epub 2016 Jun 20. Nucleic Acids Res. 2016. PMID: 27325748 Free PMC article.
-
Saccharomyces cerevisiae, a Powerful Model for Studying rRNA Modifications and Their Effects on Translation Fidelity.Int J Mol Sci. 2021 Jul 10;22(14):7419. doi: 10.3390/ijms22147419. Int J Mol Sci. 2021. PMID: 34299038 Free PMC article. Review.
-
Programmed +1 translational frameshifting in the yeast Saccharomyces cerevisiae results from disruption of translational error correction.Cold Spring Harb Symp Quant Biol. 2001;66:249-58. doi: 10.1101/sqb.2001.66.249. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762026 Review. No abstract available.
Cited by
-
Ribosomal RNA 2'-O-methylations regulate translation by impacting ribosome dynamics.Proc Natl Acad Sci U S A. 2022 Mar 22;119(12):e2117334119. doi: 10.1073/pnas.2117334119. Epub 2022 Mar 16. Proc Natl Acad Sci U S A. 2022. PMID: 35294285 Free PMC article.
-
Exonuclease-assisted enrichment and base resolution analysis of pseudouridine in single-stranded RNA.Chem Sci. 2024 Oct 21;15(45):19022-8. doi: 10.1039/d4sc03576c. Online ahead of print. Chem Sci. 2024. PMID: 39479159 Free PMC article.
-
Epitranscriptomic Signatures in lncRNAs and Their Possible Roles in Cancer.Genes (Basel). 2019 Jan 16;10(1):52. doi: 10.3390/genes10010052. Genes (Basel). 2019. PMID: 30654440 Free PMC article. Review.
-
Structural and evolutionary insights into ribosomal RNA methylation.Nat Chem Biol. 2018 Feb 14;14(3):226-235. doi: 10.1038/nchembio.2569. Nat Chem Biol. 2018. PMID: 29443970 Review.
-
Unusual C΄/D΄ motifs enable box C/D snoRNPs to modify multiple sites in the same rRNA target region.Nucleic Acids Res. 2017 Feb 28;45(4):2016-2028. doi: 10.1093/nar/gkw842. Nucleic Acids Res. 2017. PMID: 28204564 Free PMC article.
References
-
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. - PubMed
-
- Kirkwood TBL, Rosenberger RF, Galas DJ. Accuracy in Molecular Processes. London: Chapman and Hall; 1986.
-
- Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr. Opinion Struc. Biol. 2008;18:70–77. - PubMed
-
- von der Haar T, Tuite MF. Regulated translational bypass of stop codons in yeast. Trends Microbiol. 2007;15:78–86. - PubMed
-
- Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

