Thermal Stabilization of Erwinia chrysanthemi pectin methylesterase a for application in a sugar beet pulp biorefinery
- PMID: 19820151
- PMCID: PMC2786414
- DOI: 10.1128/AEM.01010-09
Thermal Stabilization of Erwinia chrysanthemi pectin methylesterase a for application in a sugar beet pulp biorefinery
Abstract
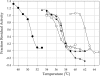
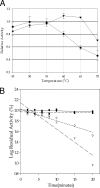
Directed evolution approaches were used to construct a thermally stabilized variant of Erwinia chrysanthemi pectin methylesterase A. The final evolved enzyme has four amino acid substitutions that together confer a T(m) value that is approximately 11 degrees C greater than that of the wild-type enzyme, while maintaining near-wild-type kinetic properties. The specific activity, with saturating substrate, of the thermally stabilized enzyme is greater than that of the wild-type enzyme when both are operating at their respective optimal temperatures, 60 degrees C and 50 degrees C. The engineered enzyme may be useful for saccharification of biomass, such as sugar beet pulp, with relatively high pectin content. In particular, the engineered enzyme is able to function in biomass up to temperatures of 65 degrees C without significant loss of activity. Specifically, the thermally stabilized enzyme facilitates the saccharification of sugar beet pulp by the commercial pectinase preparation Pectinex Ultra SPL. Added pectin methylesterase increases the initial rate of sugar production by approximately 50%.
Figures



References
-
- Beaulieu, C., M. Boccara, and F. Van Gijsegem. 1993. Pathogenic behavior of pectinase-defective Erwinia chrysanthemi mutants on different plants. Mol. Plant-Microbe Interact. 6:197-202.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

