Cryptosporidium spp. in wild, laboratory, and pet rodents in china: prevalence and molecular characterization
- PMID: 19820152
- PMCID: PMC2794099
- DOI: 10.1128/AEM.01386-09
Cryptosporidium spp. in wild, laboratory, and pet rodents in china: prevalence and molecular characterization
Abstract
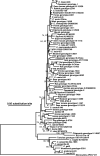
To understand the prevalence of Cryptosporidium infection in rodents in China and to assess the potential role of rodents as a source for human cryptosporidiosis, 723 specimens from 18 rodent species were collected from four provinces of China and examined between August 2007 and December 2008 by microscopy after using Sheather's sugar flotation and modified acid-fast staining. Cryptosporidium oocysts were detected in 83 specimens, with an overall prevalence of 11.5%. Phodopus sungorus, Phodopus campbelli, and Rattus tanezumi were new reported hosts of Cryptosporidium. The genotypes and subtypes of Cryptosporidium strains in microscopy-positive specimens were further identified by PCR and sequence analysis of the small subunit rRNA and the 60-kDa glycoprotein (gp60) genes. In addition to Cryptosporidium parvum, C. muris, C. andersoni, C. wrairi, ferret genotype, and mouse genotype I, four new Cryptosporidium genotypes were identified, including the hamster genotype, chipmunk genotype III, and rat genotypes II and III. Mixed Cryptosporidium species/genotypes were found in 10.8% of Cryptosporidium-positive specimens. Sequence analysis of the gp60 gene showed that C. parvum strains in pet Siberian chipmunks and hamsters were all of the subtype IIdA15G1, which was found previously in a human isolate in The Netherlands and lambs in Spain. The gp60 sequences of C. wrairi and the Cryptosporidium ferret genotype and mouse genotype I were also obtained. These findings suggest that pet rodents may be potential reservoirs of zoonotic Cryptosporidium species and subtypes.
Figures

References
-
- Alves, M., L. Xiao, V. Lemos, L. Zhou, V. Cama, M. B. da Cunha, O. Matos, and F. Antunes. 2005. Occurrence and molecular characterization of Cryptosporidium spp. in mammals and reptiles at the Lisbon zoo. Parasitol. Res. 97:108-112. - PubMed
-
- Bajer, A., M. Bednarska, A. Pawelczyk, J. M. Behnke, F. S. Gilbert, and E. Sinski. 2002. Prevalence and abundance of Cryptosporidium parvum and Giardia spp. in wild rural rodents from the Mazury Lake District region of Poland. Parasitology 125:21-34. - PubMed
-
- Bajer, A., S. Caccio, M. Bednarska, J. M. Behnke, N. J. Pieniazek, and E. Sinski. 2003. Preliminary molecular characterization of Cryptosporidium parvum isolates of wildlife rodents from Poland. J. Parasitol. 89:1053-1055. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

