Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling
- PMID: 19820169
- PMCID: PMC2775335
- DOI: 10.1073/pnas.0910177106
Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling
Abstract
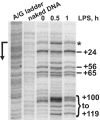
The kinetics and magnitude of cytokine gene expression are tightly regulated to elicit a balanced response to pathogens and result from integrated changes in transcription and mRNA stability. Yet, how a single microbial stimulus induces peak transcription of some genes (TNFalpha) within minutes whereas others (IP-10) require hours remains unclear. Here, we dissect activation of several lipopolysaccharide (LPS)-inducible genes in macrophages, an essential cell type mediating inflammatory response in mammals. We show that a key difference between the genes is the step of the transcription cycle at which they are regulated. Specifically, at TNFalpha, RNA Polymerase II initiates transcription in resting macrophages, but stalls near the promoter until LPS triggers rapid and transient release of the negative elongation factor (NELF) complex and productive elongation. In contrast, no NELF or polymerase is detectible near the IP-10 promoter before induction, and LPS-dependent polymerase recruitment is rate limiting for transcription. We further demonstrate that this strategy is shared by other immune mediators and is independent of the inducer and signaling pathway responsible for gene activation. Finally, as a striking example of evolutionary conservation, the Drosophila homolog of the TNFalpha gene, eiger, displayed all of the hallmarks of NELF-dependent polymerase stalling. We propose that polymerase stalling ensures the coordinated, timely activation the inflammatory gene expression program from Drosophila to mammals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–344. - PubMed
-
- Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. - PubMed
-
- Chu CQ, Field M, Feldmann M, Maini RN. Localization of tumor necrosis factor alpha in synovial tissues and at the cartilage-pannus junction in patients with rheumatoid arthritis. Arthritis Rheum. 1991;34:1125–1132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

