Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in glycine max seed coats
- PMID: 19820189
- PMCID: PMC2782299
- DOI: 10.1105/tpc.109.069856
Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in glycine max seed coats
Erratum in
- Plant Cell. 2010 May;22(5):1647
Abstract
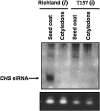
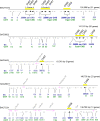
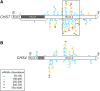
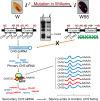
Two dominant alleles of the I locus in Glycine max silence nine chalcone synthase (CHS) genes to inhibit function of the flavonoid pathway in the seed coat. We describe here the intricacies of this naturally occurring silencing mechanism based on results from small RNA gel blots and high-throughput sequencing of small RNA populations. The two dominant alleles of the I locus encompass a 27-kb region containing two perfectly repeated and inverted clusters of three chalcone synthase genes (CHS1, CHS3, and CHS4). This structure silences the expression of all CHS genes, including CHS7 and CHS8, located on other chromosomes. The CHS short interfering RNAs (siRNAs) sequenced support a mechanism by which RNAs transcribed from the CHS inverted repeat form aberrant double-stranded RNAs that become substrates for dicer-like ribonuclease. The resulting primary siRNAs become guides that target the mRNAs of the nonlinked, highly expressed CHS7 and CHS8 genes, followed by subsequent amplification of CHS7 and CHS8 secondary siRNAs by RNA-dependent RNA polymerase. Most remarkably, this silencing mechanism occurs only in one tissue, the seed coat, as shown by the lack of CHS siRNAs in cotyledons and vegetative tissues. Thus, production of the trigger double-stranded RNA that initiates the process occurs in a specific tissue and represents an example of naturally occurring inhibition of a metabolic pathway by siRNAs in one tissue while allowing expression of the pathway and synthesis of valuable secondary metabolites in all other organs/tissues of the plant.
Figures



References
-
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.-H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36 1282–1290. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. - PubMed
-
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

