Rates of in situ transcription and splicing in large human genes
- PMID: 19820712
- PMCID: PMC2783620
- DOI: 10.1038/nsmb.1666
Rates of in situ transcription and splicing in large human genes
Abstract
Transcription and splicing must proceed over genomic distances of hundreds of kilobases in many human genes. However, the rates and mechanisms of these processes are poorly understood. We have used the compound 5,6-dichlorobenzimidazole 1-beta-D-ribofuranoside (DRB), which reversibly blocks gene transcription in vivo, combined with quantitative RT-PCR to analyze the transcription and RNA processing of several long human genes. We found that the rate of RNA polymerase II transcription over long genomic distances is about 3.8 kb min(-1) and is similar whether transcribing long introns or exon-rich regions. We also determined that co-transcriptional pre-mRNA splicing of U2-dependent introns occurs within 5-10 min of synthesis, irrespective of intron length between 1 kb and 240 kb. Similarly, U12-dependent introns were co-transcriptionally spliced within 10 min of synthesis, confirming that these introns are spliced within the nuclear compartment. These results show that the expression of large genes is unexpectedly rapid and efficient.
Figures
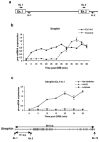
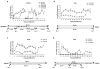
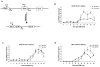
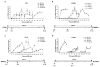
Comment in
-
Tracking rates of transcription and splicing in vivo.Nat Struct Mol Biol. 2009 Nov;16(11):1123-4. doi: 10.1038/nsmb1109-1123. Nat Struct Mol Biol. 2009. PMID: 19888309 No abstract available.
References
-
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. - PubMed
-
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. - PubMed
-
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
-
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

