Gene profiling, biomarkers and pathways characterizing HCV-related hepatocellular carcinoma
- PMID: 19821982
- PMCID: PMC2768694
- DOI: 10.1186/1479-5876-7-85
Gene profiling, biomarkers and pathways characterizing HCV-related hepatocellular carcinoma
Abstract
Background: Hepatitis C virus (HCV) infection is a major cause of hepatocellular carcinoma (HCC) worldwide. The molecular mechanisms of HCV-induced hepatocarcinogenesis are not yet fully elucidated. Besides indirect effects as tissue inflammation and regeneration, a more direct oncogenic activity of HCV can be postulated leading to an altered expression of cellular genes by early HCV viral proteins. In the present study, a comparison of gene expression patterns has been performed by microarray analysis on liver biopsies from HCV-positive HCC patients and HCV-negative controls.
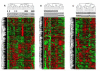
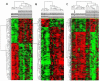
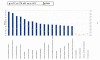
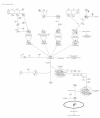
Methods: Gene expression profiling of liver tissues has been performed using a high-density microarray containing 36'000 oligos, representing 90% of the human genes. Samples were obtained from 14 patients affected by HCV-related HCC and 7 HCV-negative non-liver-cancer patients, enrolled at INT in Naples. Transcriptional profiles identified in liver biopsies from HCC nodules and paired non-adjacent non-HCC liver tissue of the same HCV-positive patients were compared to those from HCV-negative controls by the Cluster program. The pathway analysis was performed using the BRB-Array- Tools based on the "Ingenuity System Database". Significance threshold of t-test was set at 0.001.
Results: Significant differences were found between the expression patterns of several genes falling into different metabolic and inflammation/immunity pathways in HCV-related HCC tissues as well as the non-HCC counterpart compared to normal liver tissues. Only few genes were found differentially expressed between HCV-related HCC tissues and paired non-HCC counterpart.
Conclusion: In this study, informative data on the global gene expression pattern of HCV-related HCC and non-HCC counterpart, as well as on their difference with the one observed in normal liver tissues have been obtained. These results may lead to the identification of specific biomarkers relevant to develop tools for detection, diagnosis, and classification of HCV-related HCC.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

