An oligo-based microarray offers novel transcriptomic approaches for the analysis of pathogen resistance and fruit quality traits in melon (Cucumis melo L.)
- PMID: 19821986
- PMCID: PMC2767371
- DOI: 10.1186/1471-2164-10-467
An oligo-based microarray offers novel transcriptomic approaches for the analysis of pathogen resistance and fruit quality traits in melon (Cucumis melo L.)
Abstract
Background: Melon (Cucumis melo) is a horticultural specie of significant nutritional value, which belongs to the Cucurbitaceae family, whose economic importance is second only to the Solanaceae. Its small genome of approx. 450 Mb coupled to the high genetic diversity has prompted the development of genetic tools in the last decade. However, the unprecedented existence of a transcriptomic approaches in melon, highlight the importance of designing new tools for high-throughput analysis of gene expression.
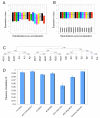
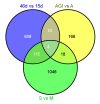
Results: We report the construction of an oligo-based microarray using a total of 17,510 unigenes derived from 33,418 high-quality melon ESTs. This chip is particularly enriched with genes that are expressed in fruit and during interaction with pathogens. Hybridizations for three independent experiments allowed the characterization of global gene expression profiles during fruit ripening, as well as in response to viral and fungal infections in plant cotyledons and roots, respectively. Microarray construction, statistical analyses and validation together with functional-enrichment analysis are presented in this study.
Conclusion: The platform validation and enrichment analyses shown in our study indicate that this oligo-based microarray is amenable for future genetic and functional genomic studies of a wide range of experimental conditions in melon.
Figures


References
-
- Garcia-Mas J, Monforte AJ, Arus P. Phylogenetic relationships among Cucumis species based on the ribosomal internal transcriber spacer sequence and microsatellite markers. Plant Syst Evol. 2004;248:191–203. doi: 10.1007/s00606-004-0170-y. - DOI
-
- Kirkbride J. Biosystematic monograph of the genus Cucumis (Cucurbitaceae) Parkway Publishers, Boone, North Carolina; 1993.
-
- Arumuganathan KEE. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. doi: 10.1007/BF02672069. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

