Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells
- PMID: 19822671
- PMCID: PMC2768842
- DOI: 10.1083/jcb.200903096
Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells
Abstract
Replicated sister chromatids are held together until mitosis by cohesin, a conserved multisubunit complex comprised of Smc1, Smc3, Scc1, and Scc3, which in vertebrate cells exists as two closely related homologues (SA1 and SA2). Here, we show that cohesin(SA1) and cohesin(SA2) are differentially required for telomere and centromere cohesion, respectively. Cells deficient in SA1 are unable to establish or maintain cohesion between sister telomeres after DNA replication in S phase. The same phenotype is observed upon depletion of the telomeric protein TIN2. In contrast, in SA2-depleted cells telomere cohesion is normal, but centromere cohesion is prematurely lost. We demonstrate that loss of telomere cohesion has dramatic consequences on chromosome morphology and function. In the absence of sister telomere cohesion, cells are unable to repair chromatid breaks and suffer sister telomere loss. Our studies elucidate the functional distinction between the Scc3 homologues in human cells and further reveal an essential role for sister telomere cohesion in genomic integrity.
Figures

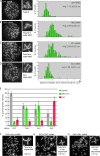
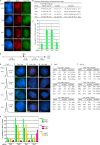
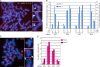
References
-
- Ancelin K., Brunori M., Bauwens S., Koering C.E., Brun C., Ricoul M., Pommier J.P., Sabatier L., Gilson E. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2.Mol. Cell. Biol. 22:3474–3487 doi:10.1128/MCB.22.10.3474-3487.2002 - DOI - PMC - PubMed
-
- Anderson D.E., Losada A., Erickson H.P., Hirano T. 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles.J. Cell Biol. 156:419–424 doi:10.1083/jcb.200111002 - DOI - PMC - PubMed
-
- Blasco M.A. 2007. The epigenetic regulation of mammalian telomeres.Nat. Rev. Genet. 8:299–309 doi:10.1038/nrg2047 - DOI - PubMed
-
- Canudas S., Houghtaling B.R., Kim J.Y., Dynek J.N., Chang W.G., Smith S. 2007. Protein requirements for sister telomere association in human cells.EMBO J. 26:4867–4878 doi:10.1038/sj.emboj.7601903 - DOI - PMC - PubMed
-
- Cesare A.J., Reddel R.R. 2008. Telomere uncapping and alternative lengthening of telomeres.Mech. Ageing Dev. 129:99–108 doi:10.1016/j.mad.2007.11.006 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

