Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors
- PMID: 19822674
- PMCID: PMC2768826
- DOI: 10.1083/jcb.200902117
Pex3 peroxisome biogenesis proteins function in peroxisome inheritance as class V myosin receptors
Abstract
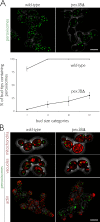
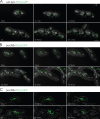
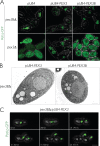
In Saccharomyces cerevisiae, peroxisomal inheritance from mother cell to bud is conducted by the class V myosin motor, Myo2p. However, homologues of S. cerevisiae Myo2p peroxisomal receptor, Inp2p, are not readily identifiable outside the Saccharomycetaceae family. Here, we demonstrate an unexpected role for Pex3 proteins in peroxisome inheritance. Both Pex3p and Pex3Bp are peroxisomal integral membrane proteins that function as peroxisomal receptors for class V myosin through direct interaction with the myosin globular tail. In cells lacking Pex3Bp, peroxisomes are preferentially retained by the mother cell, whereas most peroxisomes gather and are transferred en masse to the bud in cells overexpressing Pex3Bp or Pex3p. Our results reveal an unprecedented role for members of the Pex3 protein family in peroxisome motility and inheritance in addition to their well-established role in peroxisome biogenesis at the endoplasmic reticulum. Our results point to a temporal link between peroxisome formation and inheritance and delineate a general mechanism of peroxisome inheritance in eukaryotic cells.
Figures





References
-
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.Nucleic Acids Res. 25:3389–3402 doi:10.1093/nar/25.17.3389 - DOI - PMC - PubMed
-
- Bascom R.A., Chan H., Rachubinski R.A. 2003. Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants.Mol. Biol. Cell. 14:939–957 doi:10.1091/mbc.E02-10-0633 - DOI - PMC - PubMed
-
- Davidson R.C., Blankenship J.R., Kraus P.R., de Jesus Berrios M., Hull C.M., D'Souza C., Wang P., Heitman J. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology.Microbiology. 148:2607–2615 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

