microRNAs regulate human embryonic stem cell division
- PMID: 19823043
- PMCID: PMC2925126
- DOI: 10.4161/cc.8.22.10033
microRNAs regulate human embryonic stem cell division
Abstract
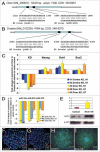
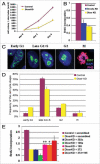
microRNAs (miRNAs) regulate numerous physiological processes such as cell division and differentiation in many tissue types including stem cells. To probe the role that miRNAs play in regulating processes relevant to embryonic stem cell biology, we used RNA interference to silence DICER and DROSHA, the two main miRNA processing enzymes. Consistent with a role for miRNAs in maintaining normal stem cell division and renewal, we found that perturbation of miRNA pathway function in human embryonic stem cells (hESCs) attenuates cell proliferation. Normal cell growth can be partially restored by introduction of the mature miRNAs miR-195 and miR-372. These miRNAs regulate two tumor suppressor genes, respectively: WEE1, which encodes a negative G2/M kinase modulator of the CycB/CDK complex and CDKN1A, which encodes p21, a CycE/CDK cyclin dependent kinase inhibitor that regulates the G1/S transition. We show that in wild-type hESCs, WEE 1 levels control the rate of hESC division, whereas p21 levels must be maintained at a low level for hESC division to proceed. These data support a model for hESC cell cycle control in which miRNAs regulate negative cell cycle modulators at two phases of the cell cycle to ensure proper replenishment of the stem cell population.
Figures


References
-
- Cheng T. Cell cycle inhibitors in normal and tumor stem cells. Oncogene. 2004;23:7256–66. - PubMed
-
- Shcherbata HR, Hatfield S, Ward EJ, Reynolds S, Fischer KA, Ruohola-Baker H. The MicroRNA pathway plays a regulatory role in stem cell division. Cell Cycle. 2006;5:172–5. - PubMed
-
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
