Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast
- PMID: 19823668
- PMCID: PMC2730531
- DOI: 10.1371/journal.pbio.1000188
Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast
Abstract
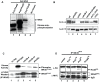
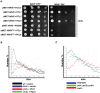
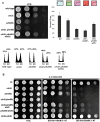
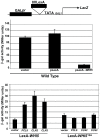
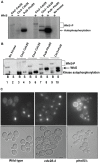
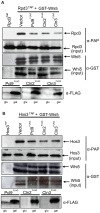
START-dependent transcription in Saccharomyces cerevisiae is regulated by two transcription factors SBF and MBF, whose activity is controlled by the binding of the repressor Whi5. Phosphorylation and removal of Whi5 by the cyclin-dependent kinase (CDK) Cln3-Cdc28 alleviates the Whi5-dependent repression on SBF and MBF, initiating entry into a new cell cycle. This Whi5-SBF/MBF transcriptional circuit is analogous to the regulatory pathway in mammalian cells that features the E2F family of G1 transcription factors and the retinoblastoma tumor suppressor protein (Rb). Here we describe genetic and biochemical evidence for the involvement of another CDK, Pcl-Pho85, in regulating G1 transcription, via phosphorylation and inhibition of Whi5. We show that a strain deleted for both PHO85 and CLN3 has a slow growth phenotype, a G1 delay, and is severely compromised for SBF-dependent reporter gene expression, yet all of these defects are alleviated by deletion of WHI5. Our biochemical and genetic tests suggest Whi5 mediates repression in part through interaction with two histone deacetylases (HDACs), Hos3 and Rpd3. In a manner analogous to cyclin D/CDK4/6, which phosphorylates Rb in mammalian cells disrupting its association with HDACs, phosphorylation by the early G1 CDKs Cln3-Cdc28 and Pcl9-Pho85 inhibits association of Whi5 with the HDACs. Contributions from multiple CDKs may provide the precision and accuracy necessary to activate G1 transcription when both internal and external cues are optimal.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Bloom J, Cross F. R. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. - PubMed
-
- Pardee A. B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. - PubMed
-
- Cross F. R. Starting the cell cycle: what's the point? Curr Opin Cell Biol. 1995;7:790–797. - PubMed
-
- Bahler J. Cell-cycle control of gene expression in budding and fission yeast. Annu Rev Genet. 2005;39:69–94. - PubMed
-
- Wittenberg C, Reed S. I. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

