RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway
- PMID: 19825650
- PMCID: PMC2902898
- DOI: 10.1093/mp/ssp006
RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway
Abstract
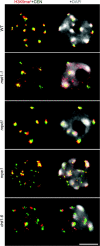
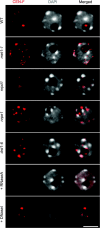
In Arabidopsis, pericentromeric repeats, retroelements, and silenced rRNA genes are assembled into heterochromatin within nuclear structures known as chromocenters. The mechanisms governing higher-order heterochromatin organization are poorly understood but 24-nt small interfering RNAs (siRNAs) are known to play key roles in heterochromatin formation. Nuclear RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2), and DICER-LIKE 3 (DCL3) are required for biogenesis of 24-nt siRNAs that associate with ARGONAUTE 4 (AGO4). Nuclear RNA polymerase V (Pol V) collaborates with DRD1 (DEFICIENT IN RNA-DEPENDENT DNA METHYLATION 1) to generate transcripts at heterochromatic loci that are hypothesized to bind to siRNA-AGO4 complexes and subsequently recruit the de-novo DNA methylation and/or histone modifying machinery. Here, we report that decondensation of the major pericentromeric repeats and depletion of the heterochromatic mark histone H3 lysine 9 dimethylation at chromocenters occurs specifically in pol V and drd1 mutants. Disruption of pericentromeric repeats condensation is coincident with transcriptional reactivation of specific classes of pericentromeric 180-bp repeats. We further demonstrate that Pol V functions independently of Pol IV, RDR2, and DCL3-mediated siRNA production to affect interphase heterochromatin organization, possibly by involving RNAs that recruit structural or chromatin-modifying proteins.
Figures




References
-
- Appelgren H, Kniola B, Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. - PubMed
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Bender J. Chromatin-based silencing mechanisms. Curr. Opin. Plant Biol. 2004;7:521–526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

