Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network
- PMID: 19826043
- PMCID: PMC2995702
- DOI: 10.1158/0008-5472.CAN-09-1567
Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network
Abstract
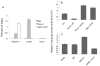
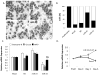
Disruption of Runx1/AML1 subnuclear localization, either by a single amino acid substitution or by a chromosomal translocation [e.g., t(8;21)], is linked to the etiology of acute myeloid leukemia (AML). Here, we show that this defect induces a select set of micro-RNAs (miR) in myeloid progenitor cells and AML patients with t(8;21). Both Runx1 and the t(8;21)-encoded AML1-ETO occupy the miR-24-23-27 locus and reciprocally control miR-24 transcription. miR-24 directly downregulates mitogen-activated protein kinase (MAPK) phosphatase-7 and enhances phosphorylation of both c-jun-NH(2)-kinase and p38 kinases. Expression of miR-24 stimulates myeloid cell growth, renders proliferation independent of interleukin-3, and blocks granulocytic differentiation. Thus, compromised Runx1 function induces a miR-dependent mechanism that, through MAPK signaling, enhances myeloid proliferation but blocks differentiation--key steps that contribute to leukemia.
Figures


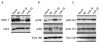
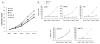
Similar articles
-
AML1/ETO proteins control POU4F1/BRN3A expression and function in t(8;21) acute myeloid leukemia.Cancer Res. 2010 May 15;70(10):3985-95. doi: 10.1158/0008-5472.CAN-09-3604. Epub 2010 May 11. Cancer Res. 2010. PMID: 20460523 Free PMC article.
-
An AML1-ETO/miR-29b-1 regulatory circuit modulates phenotypic properties of acute myeloid leukemia cells.Oncotarget. 2017 Jun 20;8(25):39994-40005. doi: 10.18632/oncotarget.18127. Oncotarget. 2017. PMID: 28611288 Free PMC article.
-
Thrombopoietin/MPL participates in initiating and maintaining RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling.Blood. 2012 Jul 26;120(4):868-79. doi: 10.1182/blood-2012-03-414649. Epub 2012 May 21. Blood. 2012. PMID: 22613795 Free PMC article.
-
Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts.Blood. 2007 Aug 1;110(3):799-805. doi: 10.1182/blood-2006-11-019265. Epub 2007 Apr 5. Blood. 2007. PMID: 17412887 Free PMC article. Review.
-
Molecular targeting of aberrant transcription factors in leukemia: strategies for RUNX1/ETO.Curr Drug Targets. 2010 Sep;11(9):1181-91. doi: 10.2174/138945010792006744. Curr Drug Targets. 2010. PMID: 20583973 Review.
Cited by
-
Targeting deregulated epigenetic control in cancer.J Cell Physiol. 2013 Nov;228(11):2103-8. doi: 10.1002/jcp.24387. J Cell Physiol. 2013. PMID: 23589100 Free PMC article. Review.
-
MicroRNA control of myelopoiesis and the differentiation block in acute myeloid leukaemia.J Cell Mol Med. 2012 May;16(5):978-87. doi: 10.1111/j.1582-4934.2011.01514.x. J Cell Mol Med. 2012. PMID: 22225649 Free PMC article. Review.
-
Versatile role of miR-24/24-1*/24-2* expression in cancer and other human diseases.Am J Transl Res. 2022 Jan 15;14(1):20-54. eCollection 2022. Am J Transl Res. 2022. PMID: 35173828 Free PMC article. Review.
-
Cardio-miRNAs and onco-miRNAs: circulating miRNA-based diagnostics for non-cancerous and cancerous diseases.Front Cell Dev Biol. 2014 Oct 16;2:61. doi: 10.3389/fcell.2014.00061. eCollection 2014. Front Cell Dev Biol. 2014. PMID: 25364765 Free PMC article. Review.
-
MiR-24 functions as a tumor suppressor in nasopharyngeal carcinoma through targeting FSCN1.J Exp Clin Cancer Res. 2015 Oct 26;34:130. doi: 10.1186/s13046-015-0242-6. J Exp Clin Cancer Res. 2015. PMID: 26503504 Free PMC article.
References
-
- Okuda T, Nishimura M, Nakao M, Fujita Y. RUNX1/AML1: a central player in hematopoiesis. Int J Hematol. 2001;74:252–7. - PubMed
-
- Downing JR. AML1/CBFbeta transcription complex: its role in normal hematopoiesis and leukemia. Leukemia. 2001;15:664–5. - PubMed
-
- Speck NA. Core binding factor and its role in normal hematopoietic development. Curr Opin Hematol. 2001;8:192–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

