Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells
- PMID: 19826477
- PMCID: PMC2757721
- DOI: 10.1371/journal.pone.0007464
Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells
Abstract
Background: Activating mutations in one allele of an oncogene (heterozygous mutations) are widely believed to be sufficient for tumorigenesis. However, mutant allele specific imbalance (MASI) has been observed in tumors and cell lines harboring mutations of oncogenes.
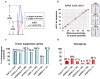
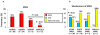
Methodology/principal findings: We determined 1) mutational status, 2) copy number gains (CNGs) and 3) relative ratio between mutant and wild type alleles of KRAS, BRAF, PIK3CA and EGFR genes by direct sequencing and quantitative PCR assay in over 400 human tumors, cell lines, and xenografts of lung, colorectal, and pancreatic cancers. Examination of a public database indicated that homozygous mutations of five oncogenes were frequent (20%) in 833 cell lines of 12 tumor types. Our data indicated two major forms of MASI: 1) MASI with CNG, either complete or partial; and 2) MASI without CNG (uniparental disomy; UPD), due to complete loss of wild type allele. MASI was a frequent event in mutant EGFR (75%) and was due mainly to CNGs, while MASI, also frequent in mutant KRAS (58%), was mainly due to UPD. Mutant: wild type allelic ratios at the genomic level were precisely maintained after transcription. KRAS mutations or CNGs were significantly associated with increased ras GTPase activity, as measured by ELISA, and the two molecular changes were synergistic. Of 237 lung adenocarcinoma tumors, the small number with both KRAS mutation and CNG were associated with shortened survival.
Conclusions: MASI is frequently present in mutant EGFR and KRAS tumor cells, and is associated with increased mutant allele transcription and gene activity. The frequent finding of mutations, CNGs and MASI occurring together in tumor cells indicates that these three genetic alterations, acting together, may have a greater role in the development or maintenance of the malignant phenotype than any individual alteration.
Conflict of interest statement
Figures

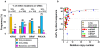



References
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. - PubMed
-
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. - PubMed
-
- Mitsudomi T, Viallet J, Mulshine JL, Linnoila RI, Minna JD, et al. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene. 1991;6:1353–1362. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

