Derived Data Storage and Exchange Workflow for Large-Scale Neuroimaging Analyses on the BIRN Grid
- PMID: 19826494
- PMCID: PMC2759340
- DOI: 10.3389/neuro.11.030.2009
Derived Data Storage and Exchange Workflow for Large-Scale Neuroimaging Analyses on the BIRN Grid
Abstract
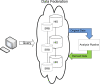
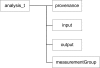
Organizing and annotating biomedical data in structured ways has gained much interest and focus in the last 30 years. Driven by decreases in digital storage costs and advances in genetics sequencing, imaging, electronic data collection, and microarray technologies, data is being collected at an ever increasing rate. The need to store and exchange data in meaningful ways in support of data analysis, hypothesis testing and future collaborative use is pervasive. Because trans-disciplinary projects rely on effective use of data from many domains, there is a genuine interest in informatics community on how best to store and combine this data while maintaining a high level of data quality and documentation. The difficulties in sharing and combining raw data become amplified after post-processing and/or data analysis in which the new dataset of interest is a function of the original data and may have been collected by multiple collaborating sites. Simple meta-data, documenting which subject and version of data were used for a particular analysis, becomes complicated by the heterogeneity of the collecting sites yet is critically important to the interpretation and reuse of derived results. This manuscript will present a case study of using the XML-Based Clinical Experiment Data Exchange (XCEDE) schema and the Human Imaging Database (HID) in the Biomedical Informatics Research Network's (BIRN) distributed environment to document and exchange derived data. The discussion includes an overview of the data structures used in both the XML and the database representations, insight into the design considerations, and the extensibility of the design to support additional analysis streams.
Keywords: BIRN; HID; MRI; XCEDE; XML; analysis; database; medical imaging.
Figures








References
-
- Arzberger P., Finholt T. A. (2002). Data and collaboratories in the biomedical community. In Report of a Panel of Experts Meeting, September 16–18, 2002, Ballston, VA
-
- Ford J. M., Roach B. J., Jorgensen K. W., Turner J. A., Brown G. G., Notestine R., Bischoff-Grethe A., Greve D., Wible C., Lauriello J., Belger A., Mueller B. A., Calhoun V., Preda A., Keator D., O'Leary D. S., Lim K. O., Glover G., Potkin S. G., Mathalon D. H. (2009). Tuning in to the voices: a multisite FMRI study of auditory hallucinations. Schizophr. Bull. 35, 58–6610.1093/schbul/sbn140 - DOI - PMC - PubMed
-
- Foster I., Vockler J., Wilde M., Zhao Y. (2003). The Virtual Data Grid: A New Model and Architecture for Data-Intensive Collaboration. silomar, CA, Proceedings of the Conference on Innovative Data Systems Research
-
- Freire J., Santos D., Silva E. (2008). Provenance for computational tasks: a survey. Comput. Sci. Eng. 10, 11–2110.1109/MCSE.2008.79 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

