Comparability of microarray data between amplified and non amplified RNA in colorectal carcinoma
- PMID: 19826639
- PMCID: PMC2760353
- DOI: 10.1155/2009/837170
Comparability of microarray data between amplified and non amplified RNA in colorectal carcinoma
Abstract
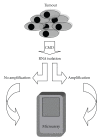
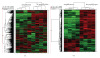
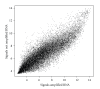
Microarray analysis reaches increasing popularity during the investigation of prognostic gene clusters in oncology. The standardisation of technical procedures will be essential to compare various datasets produced by different research groups. In several projects the amount of available tissue is limited. In such cases the preamplification of RNA might be necessary prior to microarray hybridisation. To evaluate the comparability of microarray results generated either by amplified or non amplified RNA we isolated RNA from colorectal cancer samples (stage UICC IV) following tumour tissue enrichment by macroscopic manual dissection (CMD). One part of the RNA was directly labelled and hybridised to GeneChips (HG-U133A, Affymetrix), the other part of the RNA was amplified according to the "Eberwine" protocol and was then hybridised to the microarrays. During unsupervised hierarchical clustering the samples were divided in groups regarding the RNA pre-treatment and 5.726 differentially expressed genes were identified. Using independent microarray data of 31 amplified vs. 24 non amplified RNA samples from colon carcinomas (stage UICC III) in a set of 50 predictive genes we validated the amplification bias. In conclusion microarray data resulting from different pre-processing regarding RNA pre-amplification can not be compared within one analysis.
Figures
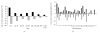
References
-
- Bueno-de-Mesquita JM, van Harten WH, Retel VP, et al. Use of 70-gene signature to predict prognosis of patients with node-negative breast cancer: a prospective community-based feasibility study (RASTER) Lancet Oncology. 2007;8(12):1079–1087. - PubMed
-
- Kim S-Y, Kim J-H, Lee H-S, et al. Meta- and gene set analysis of stomach cancer gene expression data. Molecules and Cells. 2007;24(2):200–209. - PubMed
-
- Greenawalt DM, Duong C, Smyth GK, et al. Gene expression profiling of esophageal cancer: comparative analysis of Barrett's esophagus, adenocarcinoma, and squamous cell carcinoma. International Journal of Cancer. 2007;120(9):1914–1921. - PubMed
-
- Croner RS, Peters A, Brueckl WM, et al. Microarray versus conventional prediction of lymph node metastasis in colorectal carcinoma. Cancer. 2005;104(2):395–404. - PubMed
-
- Croner RS, Foertsch T, Brueckl WM, et al. Common denominator genes that distinguish colorectal carcinoma from normal mucosa. International Journal of Colorectal Disease. 2005;20(4):353–362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

