Extraction, integration and analysis of alternative splicing and protein structure distributed information
- PMID: 19828075
- PMCID: PMC2762064
- DOI: 10.1186/1471-2105-10-S12-S15
Extraction, integration and analysis of alternative splicing and protein structure distributed information
Abstract
Background: Alternative splicing has been demonstrated to affect most of human genes; different isoforms from the same gene encode for proteins which differ for a limited number of residues, thus yielding similar structures. This suggests possible correlations between alternative splicing and protein structure. In order to support the investigation of such relationships, we have developed the Alternative Splicing and Protein Structure Scrutinizer (PASS), a Web application to automatically extract, integrate and analyze human alternative splicing and protein structure data sparsely available in the Alternative Splicing Database, Ensembl databank and Protein Data Bank. Primary data from these databases have been integrated and analyzed using the Protein Identifier Cross-Reference, BLAST, CLUSTALW and FeatureMap3D software tools.
Results: A database has been developed to store the considered primary data and the results from their analysis; a system of Perl scripts has been implemented to automatically create and update the database and analyze the integrated data; a Web interface has been implemented to make the analyses easily accessible; a database has been created to manage user accesses to the PASS Web application and store user's data and searches.
Conclusion: PASS automatically integrates data from the Alternative Splicing Database with protein structure data from the Protein Data Bank. Additionally, it comprehensively analyzes the integrated data with publicly available well-known bioinformatics tools in order to generate structural information of isoform pairs. Further analysis of such valuable information might reveal interesting relationships between alternative splicing and protein structure differences, which may be significantly associated with different functions.
Figures
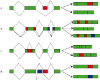

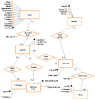



References
-
- Hiller M, Backofen R, Heymann S, Busch A, Glaesser TM, Freytag JC. Efficient prediction of alternative splice forms using protein domain homology. In Silico Biol. 2004;4:195–208. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

