Towards a systems biology approach to mammalian cell cycle: modeling the entrance into S phase of quiescent fibroblasts after serum stimulation
- PMID: 19828076
- PMCID: PMC2762065
- DOI: 10.1186/1471-2105-10-S12-S16
Towards a systems biology approach to mammalian cell cycle: modeling the entrance into S phase of quiescent fibroblasts after serum stimulation
Abstract
Background: The cell cycle is a complex process that allows eukaryotic cells to replicate chromosomal DNA and partition it into two daughter cells. A relevant regulatory step is in the G0/G1 phase, a point called the restriction (R) point where intracellular and extracellular signals are monitored and integrated.Subcellular localization of cell cycle proteins is increasingly recognized as a major factor that regulates cell cycle transitions. Nevertheless, current mathematical models of the G1/S networks of mammalian cells do not consider this aspect. Hence, there is a need for a computational model that incorporates this regulatory aspect that has a relevant role in cancer, since altered localization of key cell cycle players, notably of inhibitors of cyclin-dependent kinases, has been reported to occur in neoplastic cells and to be linked to cancer aggressiveness.
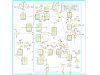
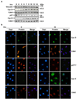
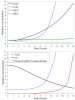
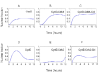
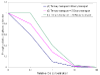
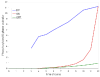
Results: The network of the model components involved in the G1 to S transition process was identified through a literature and web-based data mining and the corresponding wiring diagram of the G1 to S transition drawn with Cell Designer notation. The model has been implemented in Mathematica using Ordinary Differential Equations. Time-courses of level and of sub-cellular localization of key cell cycle players in mouse fibroblasts re-entering the cell cycle after serum starvation/re-feeding have been used to constrain network design and parameter determination. The model allows to recapitulate events from growth factor stimulation to the onset of S phase. The R point estimated by simulation is consistent with the R point experimentally determined.
Conclusion: The major element of novelty of our model of the G1 to S transition is the explicit modeling of cytoplasmic/nuclear shuttling of cyclins, cyclin-dependent kinases, their inhibitor and complexes. Sensitivity analysis of the network performance newly reveals that the biological effect brought about by Cki overexpression is strictly dependent on whether the Cki is promoting nuclear translocation of cyclin/Cdk containing complexes.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

