Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators
- PMID: 19828458
- PMCID: PMC2787342
- DOI: 10.1074/jbc.M109.065979
Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators
Abstract
Androgen-dependent transcriptional activity by the androgen receptor (AR) and its coregulators is required for male reproductive development and function. In humans and other primates, melanoma antigen gene protein-A11 (MAGE-11) is an AR selective coregulator that increases AR transcriptional activity. Here we show that the interaction between AR and MAGE-11 is mediated by AR NH(2)-terminal FXXLF motif binding to a highly conserved MAGE-11 F-box in the MAGE homology domain, and is modulated by serum stimulation of mitogen-activated protein kinase phosphorylation of MAGE-11 Ser-174. The MAGE-11-dependent increase in AR transcriptional activity is mediated by a direct interaction between MAGE-11 and transcriptional intermediary factor 2 (TIF2) through the NH(2)-terminal region of TIF2, and by a MAGE-11 FXXIF motif interaction with an F-box-like region in activation domain 1 of TIF2. The results suggest that MAGE-11 functions as a bridging factor to recruit AR coactivators through a novel FXX(L/I)F motif-F-box interaction paradigm.
Figures




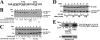



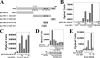



References
-
- Quigley C. A., De Bellis A., Marschke K. B., el-Awady M. K., Wilson E. M., French F. S. (1995) Endocr. Rev. 16, 271–321 - PubMed
-
- He B., Gampe R. T., Jr., Hnat A. T., Faggart J. L., Minges J. T., French F. S., Wilson E. M. (2006) J. Biol. Chem. 281, 6648–6663 - PubMed
-
- He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. (2004) Mol. Cell 16, 425–438 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

