Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants
- PMID: 19828610
- PMCID: PMC2798406
- DOI: 10.1128/JVI.02199-08
Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants
Abstract
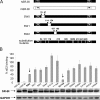
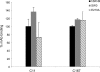
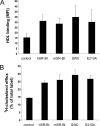
Scavenger receptor class B type I (SR-BI) is an essential receptor for hepatitis C virus (HCV) and a cell surface high-density-lipoprotein (HDL) receptor. The mechanism of SR-BI-mediated HCV entry, however, is not clearly understood, and the specific protein determinants required for the recognition of the virus envelope are not known. HCV infection is strictly linked to lipoprotein metabolism, and HCV virions may initially interact with SR-BI through associated lipoproteins before subsequent direct interactions of the viral glycoproteins with SR-BI occur. The kinetics of inhibition of cell culture-derived HCV (HCVcc) infection with an anti-SR-BI monoclonal antibody imply that the recognition of SR-BI by HCV is an early event of the infection process. Swapping and single-substitution mutants between mouse and human SR-BI sequences showed reduced binding to the recombinant soluble E2 (sE2) envelope glycoprotein, thus suggesting that the SR-BI interaction with the HCV envelope is likely to involve species-specific protein elements. Most importantly, SR-BI mutants defective for sE2 binding, although retaining wild-type activity for receptor oligomerization and binding to the physiological ligand HDL, were impaired in their ability to fully restore HCVcc infectivity when transduced into an SR-BI-knocked-down Huh-7.5 cell line. These findings suggest a specific and direct role for the identified residues in binding HCV and mediating virus entry. Moreover, the observation that different regions of SR-BI are involved in HCV and HDL binding supports the hypothesis that new therapeutic strategies aimed at interfering with virus/SR-BI recognition are feasible.
Figures




Similar articles
-
High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein.J Virol. 2007 Aug;81(15):8063-71. doi: 10.1128/JVI.00193-07. Epub 2007 May 16. J Virol. 2007. PMID: 17507483 Free PMC article.
-
High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI.J Biol Chem. 2006 Jul 7;281(27):18285-95. doi: 10.1074/jbc.M602706200. Epub 2006 May 4. J Biol Chem. 2006. PMID: 16675450
-
Single amino acid mutation of SR-BI decreases infectivity of hepatitis C virus derived from cell culture in a cell culture model.World J Gastroenterol. 2017 Jul 28;23(28):5158-5166. doi: 10.3748/wjg.v23.i28.5158. World J Gastroenterol. 2017. PMID: 28811710 Free PMC article.
-
Scavenger receptor class B type I and the hypervariable region-1 of hepatitis C virus in cell entry and neutralisation.Expert Rev Mol Med. 2011 Apr 14;13:e13. doi: 10.1017/S1462399411001785. Expert Rev Mol Med. 2011. PMID: 21489334 Review.
-
Hepatitis C virus entry: potential receptors and their biological functions.J Gen Virol. 2006 May;87(Pt 5):1075-1084. doi: 10.1099/vir.0.81646-0. J Gen Virol. 2006. PMID: 16603507 Review.
Cited by
-
Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies.PLoS Pathog. 2013 Mar;9(3):e1003244. doi: 10.1371/journal.ppat.1003244. Epub 2013 Mar 21. PLoS Pathog. 2013. PMID: 23555257 Free PMC article.
-
Lipoprotein Receptors Redundantly Participate in Entry of Hepatitis C Virus.PLoS Pathog. 2016 May 6;12(5):e1005610. doi: 10.1371/journal.ppat.1005610. eCollection 2016 May. PLoS Pathog. 2016. PMID: 27152966 Free PMC article.
-
Comparison of the inhibitory and stimulatory effects of Core and NS3 candidate HCV vaccines on the cellular immune response.Am J Clin Exp Immunol. 2023 Dec 15;12(6):153-163. eCollection 2023. Am J Clin Exp Immunol. 2023. PMID: 38187363 Free PMC article.
-
Molecular consequences of SARS-CoV-2 liver tropism.Nat Metab. 2022 Mar;4(3):310-319. doi: 10.1038/s42255-022-00552-6. Epub 2022 Mar 28. Nat Metab. 2022. PMID: 35347318 Free PMC article.
-
Impact of intra- and interspecies variation of occludin on its function as coreceptor for authentic hepatitis C virus particles.J Virol. 2011 Aug;85(15):7613-21. doi: 10.1128/JVI.00212-11. Epub 2011 Jun 1. J Virol. 2011. PMID: 21632765 Free PMC article.
References
-
- Acton, S., A. Rigotti, K. T. Landschulz, S. Xu, H. H. Hobbs, and M. Krieger. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271:518-520. - PubMed
-
- Allander, T., X. Forns, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology 277:358-367. - PubMed
-
- Barth, H., R. Cerino, M. Arcuri, M. Hoffmann, P. Schurmann, M. I. Adah, B. Gissler, X. Zhao, V. Ghisetti, B. Lavezzo, H. E. Blum, F. von Weizsacker, A. Vitelli, E. Scarselli, and T. F. Baumert. 2005. Scavenger receptor class B type I and hepatitis C virus infection of primary tupaia hepatocytes. J. Virol. 79:5774-5785. - PMC - PubMed
-
- Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

