Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity of HRV species C
- PMID: 19828751
- PMCID: PMC2786677
- DOI: 10.1128/JCM.00993-09
Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity of HRV species C
Abstract
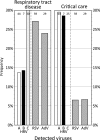
Rhinovirus infections are the most common cause of viral illness in humans, and there is increasing evidence of their etiological role in severe acute respiratory tract infections (ARTIs). Human rhinoviruses (HRVs) are classified into two species, species A and B, which contain over 100 serotypes, and a recently discovered genetically heterogeneous third species (HRV species C). To investigate their diversity and population turnover, screening for the detection and the genetic characterization of HRV variants in diagnostic respiratory samples was performed by using nested primers for the efficient amplification of the VP4-VP2 region of HRV (and enterovirus) species and serotype identification. HRV species A, B, and C variants were detected in 14%, 1.8%, and 6.8%, respectively, of 456 diagnostic respiratory samples from 345 subjects (6 samples also contained enteroviruses), predominantly among children under age 10 years. HRV species A and B variants were remarkably heterogeneous, with 22 and 6 different serotypes, respectively, detected among 73 positive samples. Similarly, by using a pairwise distance threshold of 0.1, species C variants occurring worldwide were provisionally assigned to 47 different types, of which 15 were present among samples from Edinburgh, United Kingdom. There was a rapid turnover of variants, with only 5 of 43 serotypes detected during both sampling periods. By using divergence thresholds and phylogenetic analysis, several species A and C variants could provisionally be assigned to new types. An initial investigation of the clinical differences between rhinovirus species found HRV species C to be nearly twice as frequently associated with ARTIs than other rhinovirus species, which matches the frequencies of detection of respiratory syncytial virus. The study demonstrates the extraordinary genetic diversity of HRVs, their rapid population turnover, and their extensive involvement in childhood respiratory disease.
Figures




References
-
- Blomqvist, S., M. Roivainen, T. Puhakka, M. Kleemola, and T. Hovi. 2002. Virological and serological analysis of rhinovirus infections during the first two years of life in a cohort of children. J. Med. Virol. 66:263-268. - PubMed
-
- Briese, T., N. Renwick, M. Venter, R. G. Jarman, D. Ghosh, S. Kondgen, S. K. Shrestha, A. M. Hoegh, I. Casas, E. V. Adjogoua, C. Akoua-Koffi, K. S. Myint, D. T. Williams, G. Chidlow, R. van den Berg, C. Calvo, O. Koch, G. Palacios, V. Kapoor, J. Villari, S. R. Dominguez, K. V. Holmes, G. Harnett, D. Smith, J. S. Mackenzie, H. Ellerbrok, B. Schweiger, K. Schonning, M. S. Chadha, F. H. Leendertz, A. C. Mishra, R. V. Gibbons, E. C. Holmes, and W. I. Lipkin. 2008. Global distribution of novel rhinovirus genotype. Emerg. Infect. Dis. 14:944-947. - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

