Human DNA methylomes at base resolution show widespread epigenomic differences
- PMID: 19829295
- PMCID: PMC2857523
- DOI: 10.1038/nature08514
Human DNA methylomes at base resolution show widespread epigenomic differences
Abstract
DNA cytosine methylation is a central epigenetic modification that has essential roles in cellular processes including genome regulation, development and disease. Here we present the first genome-wide, single-base-resolution maps of methylated cytosines in a mammalian genome, from both human embryonic stem cells and fetal fibroblasts, along with comparative analysis of messenger RNA and small RNA components of the transcriptome, several histone modifications, and sites of DNA-protein interaction for several key regulatory factors. Widespread differences were identified in the composition and patterning of cytosine methylation between the two genomes. Nearly one-quarter of all methylation identified in embryonic stem cells was in a non-CG context, suggesting that embryonic stem cells may use different methylation mechanisms to affect gene regulation. Methylation in non-CG contexts showed enrichment in gene bodies and depletion in protein binding sites and enhancers. Non-CG methylation disappeared upon induced differentiation of the embryonic stem cells, and was restored in induced pluripotent stem cells. We identified hundreds of differentially methylated regions proximal to genes involved in pluripotency and differentiation, and widespread reduced methylation levels in fibroblasts associated with lower transcriptional activity. These reference epigenomes provide a foundation for future studies exploring this key epigenetic modification in human disease and development.
Figures
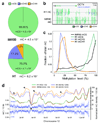

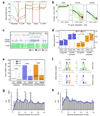
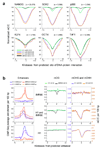

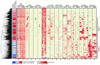
Comment in
-
Epigenomics: Methylation matters.Nature. 2009 Nov 19;462(7271):296-7. doi: 10.1038/462296a. Nature. 2009. PMID: 19924205 No abstract available.
References
-
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. - PubMed
-
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. - PubMed
-
- Bestor TH. The DNA methyltransferases of mammals. Human Molecular Genetics. 2000;9:2395–2402. - PubMed
-
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
-
- Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

