DUX4c is up-regulated in FSHD. It induces the MYF5 protein and human myoblast proliferation
- PMID: 19829708
- PMCID: PMC2759506
- DOI: 10.1371/journal.pone.0007482
DUX4c is up-regulated in FSHD. It induces the MYF5 protein and human myoblast proliferation
Abstract
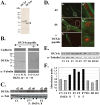
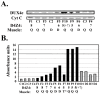
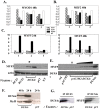
Facioscapulohumeral muscular dystrophy (FSHD) is a dominant disease linked to contractions of the D4Z4 repeat array in 4q35. We have previously identified a double homeobox gene (DUX4) within each D4Z4 unit that encodes a transcription factor expressed in FSHD but not control myoblasts. DUX4 and its target genes contribute to the global dysregulation of gene expression observed in FSHD. We have now characterized the homologous DUX4c gene mapped 42 kb centromeric of the D4Z4 repeat array. It encodes a 47-kDa protein with a double homeodomain identical to DUX4 but divergent in the carboxyl-terminal region. DUX4c was detected in primary myoblast extracts by Western blot with a specific antiserum, and was induced upon differentiation. The protein was increased about 2-fold in FSHD versus control myotubes but reached 2-10-fold induction in FSHD muscle biopsies. We have shown by Western blot and by a DNA-binding assay that DUX4c over-expression induced the MYF5 myogenic regulator and its DNA-binding activity. DUX4c might stabilize the MYF5 protein as we detected their interaction by co-immunoprecipitation. In keeping with the known role of Myf5 in myoblast accumulation during mouse muscle regeneration DUX4c over-expression activated proliferation of human primary myoblasts and inhibited their differentiation. Altogether, these results suggested that DUX4c could be involved in muscle regeneration and that changes in its expression could contribute to the FSHD pathology.
Conflict of interest statement
Figures



References
-
- Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet. 1994;3:1287–1295. - PubMed
-
- Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. - PubMed
-
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. - PubMed
-
- Lunt PW, Jardine PE, Koch MC, Maynard J, Osborn M, et al. Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD) [published erratum appears in Hum Mol Genet 1995 Jul;4(7):1243-4]. Hum Mol Genet. 1995;4:951–958. - PubMed
-
- Ricci E, Galluzzi G, Deidda G, Cacurri S, Colantoni L, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol. 1999;45:751–757. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

