The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA
- PMID: 19833920
- PMCID: PMC3763470
- DOI: 10.1126/science.1179700
The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA
Abstract
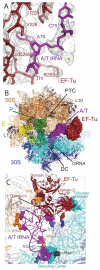
The ribosome selects a correct transfer RNA (tRNA) for each amino acid added to the polypeptide chain, as directed by messenger RNA. Aminoacyl-tRNA is delivered to the ribosome by elongation factor Tu (EF-Tu), which hydrolyzes guanosine triphosphate (GTP) and releases tRNA in response to codon recognition. The signaling pathway that leads to GTP hydrolysis upon codon recognition is critical to accurate decoding. Here we present the crystal structure of the ribosome complexed with EF-Tu and aminoacyl-tRNA, refined to 3.6 angstrom resolution. The structure reveals details of the tRNA distortion that allows aminoacyl-tRNA to interact simultaneously with the decoding center of the 30S subunit and EF-Tu at the factor binding site. A series of conformational changes in EF-Tu and aminoacyl-tRNA suggests a communication pathway between the decoding center and the guanosine triphosphatase center of EF-Tu.
Figures



Comment in
-
Biochemistry. Leaps in translational elongation.Science. 2009 Oct 30;326(5953):677-8. doi: 10.1126/science.1181511. Epub 2009 Oct 15. Science. 2009. PMID: 19833922 No abstract available.
References
-
- Rodnina MV, Pape T, Fricke R, Kuhn L, Wintermeyer W. J Biol Chem. 1996;271:646. - PubMed
-
- Ogle JM, et al. Science. 2001;292:897. - PubMed
-
- Gromadski KB, Rodnina MV. Mol Cell. 2004;13:191. - PubMed
-
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Cell. 2002;111:721. - PubMed
-
- Xia T, et al. Biochemistry. 1998;37:14719. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

