Histone H1 binding is inhibited by histone variant H3.3
- PMID: 19834459
- PMCID: PMC2790488
- DOI: 10.1038/emboj.2009.301
Histone H1 binding is inhibited by histone variant H3.3
Abstract
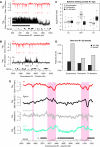
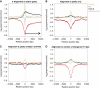
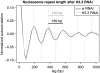
Linker histones are involved in the formation of higher-order chromatin structure and the regulation of specific genes, yet it remains unclear what their principal binding determinants are. We generated a genome-wide high-resolution binding map for linker histone H1 in Drosophila cells, using DamID. H1 binds at similar levels across much of the genome, both in classic euchromatin and heterochromatin. Strikingly, there are pronounced dips of low H1 occupancy around transcription start sites for active genes and at many distant cis-regulatory sites. H1 dips are not due to lack of nucleosomes; rather, all regions with low binding of H1 show enrichment of the histone variant H3.3. Knockdown of H3.3 causes H1 levels to increase at these sites, with a concomitant increase in nucleosome repeat length. These changes are independent of transcriptional changes. Our results show that the H3.3 protein counteracts association of H1, providing a mechanism to keep diverse genomic sites in an open chromatin conformation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Ahmad K, Henikoff S (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Chiu N, Baserga R, Furth JJ (1977) Composition and template activity of chromatin fractionated by isoelectric focusing. Biochemistry 16: 4796–4802 - PubMed
-
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH (2006) Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell 11: 775–789 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

