Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly
- PMID: 19834536
- PMCID: PMC2752994
- DOI: 10.1371/journal.pgen.1000682
Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly
Abstract
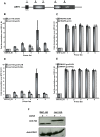
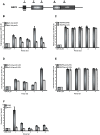
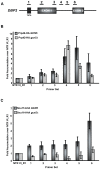
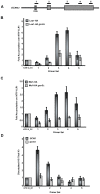
In the last several years, a number of studies have shown that spliceosome assembly and splicing catalysis can occur co-transcriptionally. However, it has been unclear which specific transcription factors play key roles in coupling splicing to transcription and the mechanisms through which they act. Here we report the discovery that Gcn5, which encodes the histone acetyltransferase (HAT) activity of the SAGA complex, has genetic interactions with the genes encoding the heterodimeric U2 snRNP proteins Msl1 and Lea1. These interactions are dependent upon the HAT activity of Gcn5, suggesting a functional relationship between Gcn5 HAT activity and Msl1/Lea1 function. To understand the relationship between Gcn5 and Msl1/Lea1, we carried out an analysis of Gcn5's role in co-transcriptional recruitment of Msl1 and Lea1 to pre-mRNA and found that Gcn5 HAT activity is required for co-transcriptional recruitment of the U2 snRNP (and subsequent snRNP) components to the branchpoint, while it is not required for U1 recruitment. Although previous studies suggest that transcription elongation can alter co-transcriptional pre-mRNA splicing, we do not observe evidence of defective transcription elongation for these genes in the absence of Gcn5, while Gcn5-dependent histone acetylation is enriched in the promoter regions. Unexpectedly, we also observe Msl1 enrichment in the promoter region for wild-type cells and cells lacking Gcn5, indicating that Msl1 recruitment during active transcription can occur independently of its association at the branchpoint region. These results demonstrate a novel role for acetylation by SAGA in co-transcriptional recruitment of the U2 snRNP and recognition of the intron branchpoint.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. - PubMed
-
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1998;2:754–65. - PubMed
-
- Beyer AL, Osheim YN. Visualization of RNA transcription and processing. Semin Cell Biol. 1991;2:131–40. - PubMed
-
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

