A novel mechanism of transposon-mediated gene activation
- PMID: 19834539
- PMCID: PMC2753651
- DOI: 10.1371/journal.pgen.1000689
A novel mechanism of transposon-mediated gene activation
Abstract
Transposable Insertion Sequences (IS elements) have been shown to provide various benefits to their hosts via gene activation or inactivation under stress conditions by appropriately inserting into specific chromosomal sites. Activation is usually due to derepression or introduction of a complete or partial promoter located within the element. Here we define a novel mechanism of gene activation by the transposon IS5 in Escherichia coli. The glycerol utilization operon, glpFK, that is silent in the absence of the cAMP-Crp complex, is activated by IS5 when inserted upstream of its promoter. High-level expression is nearly constitutive, only mildly dependent on glycerol, glucose, GlpR, and Crp, and allows growth at a rate similar to or more rapid than that of wild-type cells. Expression is from the glpFK promoter and dependent on (1) the DNA phase, (2) integration host factor (IHF), and (3) a short region at the 3' end of IS5 harboring a permanent bend and an IHF binding site. The lacZYA operon is also subject to such activation in the absence of Crp. Thus, we have defined a novel mechanism of gene activation involving transposon insertion that may be generally applicable to many organisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

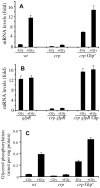
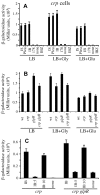

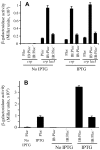
References
-
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995.
-
- Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G, Hengge-Aronis R, editors. Bacterial Stress Response. Washington, DC: ASM Press; 2000. pp. 161–178.
-
- Chandler M, Mahillon J. Insertion sequences revised. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 305–366.
-
- Galas DJ, Chandler M. Bacterial insertion sequences. In: Berg D, Howe M, editors. Mobile DNA. Washington DC: American Society for Microbiology; 1989. pp. 109–162.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

