Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses
- PMID: 19834549
- PMCID: PMC2757677
- DOI: 10.1371/journal.ppat.1000627
Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses
Abstract
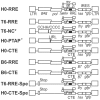
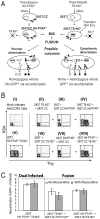
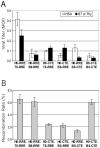
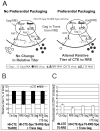
Once transcribed, the nascent full-length RNA of HIV-1 must travel to the appropriate host cell sites to be translated or to find a partner RNA for copackaging to form newly generated viruses. In this report, we sought to delineate the location where HIV-1 RNA initiates dimerization and the influence of the RNA transport pathway used by the virus on downstream events essential to viral replication. Using a cell-fusion-dependent recombination assay, we demonstrate that the two RNAs destined for copackaging into the same virion select each other mostly within the cytoplasm. Moreover, by manipulating the RNA export element in the viral genome, we show that the export pathway taken is important for the ability of RNA molecules derived from two viruses to interact and be copackaged. These results further illustrate that at the point of dimerization the two main cellular export pathways are partially distinct. Lastly, by providing Gag in trans, we have demonstrated that Gag is able to package RNA from either export pathway, irrespective of the transport pathway used by the gag mRNA. These findings provide unique insights into the process of RNA export in general, and more specifically, of HIV-1 genomic RNA trafficking.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Hammarskjold ML. Regulation of retroviral RNA export. Semin Cell Dev Biol. 1997;8:83–90. - PubMed
-
- Swanson CM, Malim MH. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic. 2006;7:1440–1450. - PubMed
-
- Rodriguez MS, Dargemont C, Stutz F. Nuclear export of RNA. Biol Cell. 2004;96:639–655. - PubMed
-
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

