The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module
- PMID: 19835885
- PMCID: PMC2806952
- DOI: 10.1016/j.jmb.2009.10.010
The central portion of factor H (modules 10-15) is compact and contains a structurally deviant CCP module
Abstract
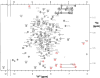
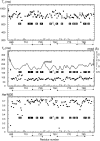
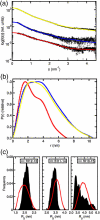
The first eight and the last two of 20 complement control protein (CCP) modules within complement factor H (fH) encompass binding sites for C3b and polyanionic carbohydrates. These binding sites cooperate self-surface selectively to prevent C3b amplification, thus minimising complement-mediated damage to host. Intervening fH CCPs, apparently devoid of such recognition sites, are proposed to play a structural role. One suggestion is that the generally small CCPs 10-15, connected by longer-than-average linkers, act as a flexible tether between the two functional ends of fH; another is that the long linkers induce a 180 degrees bend in the middle of fH. To test these hypotheses, we determined the NMR-derived structure of fH12-13 consisting of module 12, shown here to have an archetypal CCP structure, and module 13, which is uniquely short and features a laterally protruding helix-like insertion that contributes to a prominent electropositive patch. The unusually long fH12-13 linker is not flexible. It packs between the two CCPs that are not folded back on each other but form a shallow vee shape; analytical ultracentrifugation and X-ray scattering supported this finding. These two techniques additionally indicate that flanking modules (within fH11-14 and fH10-15) are at least as rigid and tilted relative to neighbours as are CCPs 12 and 13 with respect to one another. Tilts between successive modules are not unidirectional; their principal axes trace a zigzag path. In one of two arrangements for CCPs 10-15 that fit well with scattering data, CCP 14 is folded back onto CCP 13. In conclusion, fH10-15 forms neither a flexible tether nor a smooth bend. Rather, it is compact and has embedded within it a CCP module (CCP 13) that appears to be highly specialised given both its deviant structure and its striking surface charge distribution. A passive, purely structural role for this central portion of fH is unlikely.
Figures





References
-
- Walport M.J. Complement. First of two parts. N. Engl. J. Med. 2001;344:1058–1066. - PubMed
-
- Walport M.J. Complement. Second of two parts. N. Engl. J. Med. 2001;344:1140–1144. - PubMed
-
- Muller-Eberhard H.J. Transmembrane channel-formation by five complement proteins. Biochem. Soc. Symp. 1985;50:235–246. - PubMed
-
- Liszewski M.K., Farries T.C., Lublin D.M., Rooney I.A., Atkinson J.P. Control of the complement system. Adv. Immunol. 1996;61:201–283. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

