Structure of IL-33 and its interaction with the ST2 and IL-1RAcP receptors--insight into heterotrimeric IL-1 signaling complexes
- PMID: 19836339
- PMCID: PMC2766095
- DOI: 10.1016/j.str.2009.08.009
Structure of IL-33 and its interaction with the ST2 and IL-1RAcP receptors--insight into heterotrimeric IL-1 signaling complexes
Abstract
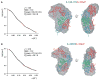
Members of the interleukin-1 (IL-1) family of cytokines play major roles in host defense and immune system regulation in infectious and inflammatory diseases. IL-1 cytokines trigger a biological response in effector cells by assembling a heterotrimeric signaling complex with two IL-1 receptor chains, a high-affinity primary receptor and a low-affinity coreceptor. To gain insights into the signaling mechanism of the novel IL-1-like cytokine IL-33, we first solved its solution structure and then performed a detailed biochemical and structural characterization of the interaction between IL-33, its primary receptor ST2, and the coreceptor IL-1RAcP. Using nuclear magnetic resonance data, we obtained a model of the IL-33/ST2 complex in solution that is validated by small-angle X-ray scattering (SAXS) data and is similar to the IL-1beta/IL-1R1 complex. We extended our SAXS analysis to the IL-33/ST2/IL-1RAcP and IL-1beta/IL-1R1/IL-1RAcP complexes and propose a general model of the molecular architecture of IL-1 ternary signaling complexes.
Figures






References
-
- Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. - PubMed
-
- Boraschi D, Bossu P, Ruggiero P, Tagliabue A, Bertini R, Macchia G, Gasbarro C, Pellegrini L, Melillo G, Ulisse E, et al. Mapping of receptor binding sites on IL-1 beta by reconstruction of IL-1ra-like domains. J Immunol. 1995;155:4719–4725. - PubMed
-
- Boraschi D, Tagliabue A. The interleukin-1 receptor family. Vitam Horm. 2006;74:229–254. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

