Adenylate-forming enzymes
- PMID: 19836944
- PMCID: PMC3313645
- DOI: 10.1016/j.sbi.2009.09.004
Adenylate-forming enzymes
Abstract
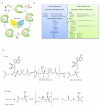
Thioesters, amides, and esters are common chemical building blocks in a wide array of natural products. The formation of these bonds can be catalyzed in a variety of ways. For chemists, the use of an activating group is a common strategy and adenylate enzymes are exemplars of this approach. Adenylating enzymes activate the otherwise unreactive carboxylic acid by transforming the normal hydroxyl leaving group into adenosine monophosphate. Recently there have been a number of studies of such enzymes and in this review we suggest a new classification scheme. The review highlights the diversity in enzyme fold, active site architecture, and metal coordination that has evolved to catalyze this particular reaction.
Figures


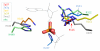
Similar articles
-
Repurposing the 3-Isocyanobutanoic Acid Adenylation Enzyme SfaB for Versatile Amidation and Thioesterification.Angew Chem Int Ed Engl. 2021 Jan 25;60(4):2030-2035. doi: 10.1002/anie.202010042. Epub 2020 Nov 24. Angew Chem Int Ed Engl. 2021. PMID: 33026145
-
The adenylate energy charge in the study of enzymes in vitro.Methods Enzymol. 1979;55:229-35. doi: 10.1016/0076-6879(79)55027-7. Methods Enzymol. 1979. PMID: 459842 No abstract available.
-
Enzymatic Strategies for the Biosynthesis of N-Acyl Amino Acid Amides.Chembiochem. 2024 Feb 16;25(4):e202300672. doi: 10.1002/cbic.202300672. Epub 2024 Jan 5. Chembiochem. 2024. PMID: 38051126 Review.
-
Enzyme millisecond conformational dynamics do not catalyze the chemical step.Proc Natl Acad Sci U S A. 2009 Oct 13;106(41):17359-64. doi: 10.1073/pnas.0909150106. Epub 2009 Sep 25. Proc Natl Acad Sci U S A. 2009. PMID: 19805169 Free PMC article.
-
Emerging enzymes for ATP regeneration in biocatalytic processes.Chembiochem. 2015 Feb 9;16(3):380-6. doi: 10.1002/cbic.201402550. Epub 2015 Jan 23. Chembiochem. 2015. PMID: 25619338 Review.
Cited by
-
In vitro reconstitution of indolmycin biosynthesis reveals the molecular basis of oxazolinone assembly.Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):2717-22. doi: 10.1073/pnas.1419964112. Epub 2015 Feb 17. Proc Natl Acad Sci U S A. 2015. PMID: 25730866 Free PMC article.
-
The catalytic mechanism of the hotdog-fold enzyme superfamily 4-hydroxybenzoyl-CoA thioesterase from Arthrobacter sp. strain SU.Biochemistry. 2012 Sep 4;51(35):7000-16. doi: 10.1021/bi301059m. Epub 2012 Aug 20. Biochemistry. 2012. PMID: 22873756 Free PMC article.
-
Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis.Plant Cell. 2012 Nov;24(11):4525-38. doi: 10.1105/tpc.112.102921. Epub 2012 Nov 6. Plant Cell. 2012. PMID: 23136372 Free PMC article.
-
Improved Synthesis of Biotinol-5'-AMP: Implications for Antibacterial Discovery.ACS Med Chem Lett. 2014 Dec 11;6(2):216-20. doi: 10.1021/ml500475n. eCollection 2015 Feb 12. ACS Med Chem Lett. 2014. PMID: 25699152 Free PMC article.
-
Cloning and Immunosuppressive Properties of an Acyl-Activating Enzyme from the Venom Apparatus of Tetrastichus brontispae (Hymenoptera: Eulophidae).Toxins (Basel). 2019 Nov 18;11(11):672. doi: 10.3390/toxins11110672. Toxins (Basel). 2019. PMID: 31752154 Free PMC article.
References
-
- Du L, He Y, Luo Y. Crystal structure and enantiomer selection by D-alanyl carrier protein ligase DltA from Bacillus cereus. Biochemistry. 2008;47:11473–11480. - PubMed
-
- Fulda M, Heinz E, Wolter FP. The fadD gene of Escherichia coli K12 is located close to rnd at 39.6 min of the chromosomal map and is a new member of the AMP-binding protein family. Mol Gen Genet. 1994;242:241–249. - PubMed
-
- Jogl G, Tong L. Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry. 2004;43:1425–1431. - PubMed
-
- Schmelz S, Kadi N, McMahon SA, Song LJ, Oves-Costales D, Oke M, Liu HT, Johnson KA, Carter LG, Botting CH, et al. AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis. Nature Chemical Biology. 2009;5:174–182. - PMC - PubMed
-
This paper reports the chemical and structural characterization of the first class III member of adenylate-forming enzyme. The enzyme makes AMP-citrate which is then condensed with L-serine to make a citrate ester. The paper highlights the similarity in both chemistry and structure with kinases and adenylate-forming enzymes.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

