Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging
- PMID: 19836954
- PMCID: PMC3663713
- DOI: 10.1016/j.tcb.2009.09.003
Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging
Abstract
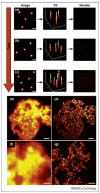
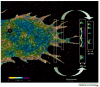
Photoactivatable fluorescent proteins (PA-FPs) are molecules that switch to a new fluorescent state in response to activation to generate a high level of contrast. Over the past eight years, several types of PA-FPs have been developed. The PA-FPs fluoresce green or red, or convert from green to red in response to activating light. Others reversibly switch between 'off' and 'on' in response to light. The optical "highlighting" capability of PA-FPs has led to the rise of novel imaging techniques providing important new biological insights. These range from in cellulo pulse-chase labeling for tracking subpopulations of cells, organelles or proteins under physiological settings, to super-resolution imaging of single molecules for determining intracellular protein distributions at nanometer precision. This review surveys the expanding array of PA-FPs, including their advantages and disadvantages, and highlights their use in novel imaging methodologies.
Figures





References
-
- Tsien RY. The green fluorescent protein. Ann Rev Biochem. 1998;67:509–544. - PubMed
-
- Lukyanov KA, et al. Innovation: photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. - PubMed
-
- Lippincott-Schwartz J, Patterson GH. Fluorescent proteins for photoactivation experiments. Methods Cell Biol. 2008;85:45–61. - PubMed
-
- Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–943. - PubMed
-
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

