GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding
- PMID: 19837035
- PMCID: PMC2779841
- DOI: 10.1016/j.cell.2009.07.052
GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding
Abstract
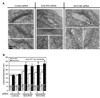
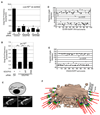
Golgi membranes, from yeast to humans, are uniquely enriched in phosphatidylinositol-4-phosphate (PtdIns(4)P), although the role of this lipid remains poorly understood. Using a proteomic lipid-binding screen, we identify the Golgi protein GOLPH3 (also called GPP34, GMx33, MIDAS, or yeast Vps74p) as a PtdIns(4)P-binding protein that depends on PtdIns(4)P for its Golgi localization. We further show that GOLPH3 binds the unconventional myosin MYO18A, thus connecting the Golgi to F-actin. We demonstrate that this linkage is necessary for normal Golgi trafficking and morphology. The evidence suggests that GOLPH3 binds to PtdIns(4)P-rich trans-Golgi membranes and MYO18A conveying a tensile force required for efficient tubule and vesicle formation. Consequently, this tensile force stretches the Golgi into the extended ribbon observed by fluorescence microscopy and the familiar flattened form observed by electron microscopy.
Figures
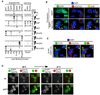

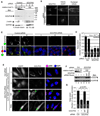
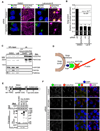
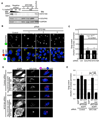
References
-
- Allan VJ, Thompson HM, McNiven MA. Motoring around the Golgi. Nat. Cell Biol. 2002;4:E236–E242. - PubMed
-
- Barr FA, Warren G. Disassembly and reassembly of the Golgi apparatus. Semin. Cell Dev. Biol. 1996;7:505–510.
-
- Bell AW, Ward MA, Blackstock WP, Freeman HNM, Choudhary JS, Lewis AP, Chotai D, Fazel A, Gushue JN, Paiement J, Palcy S, Chevet E, Lafrenière-Roula M, Solari R, Thomas DY, Rowley A, Bergeron JJM. Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 2001;276:5152–5165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

