Substrate-induced assembly of Methanococcoides burtonii D-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers
- PMID: 19837658
- PMCID: PMC2797158
- DOI: 10.1074/jbc.M109.050989
Substrate-induced assembly of Methanococcoides burtonii D-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers
Abstract
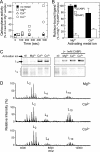
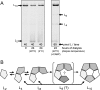
Like many enzymes, the biogenesis of the multi-subunit CO(2)-fixing enzyme ribulose-1,5-bisphosphate (RuBP) carboxylase/oxygenase (Rubisco) in different organisms requires molecular chaperones. When expressed in Escherichia coli, the large (L) subunits of the Rubisco from the archaeabacterium Methanococcoides burtonii assemble into functional dimers (L(2)). However, further assembly into pentamers of L(2) (L(10)) occurs when expressed in tobacco chloroplasts or E. coli producing RuBP. In vitro analyses indicate that the sequential assembly of L(2) into L(10) (via detectable L(4) and L(6) intermediates) occurs without chaperone involvement and is stimulated by protein rearrangements associated with either the binding of substrate RuBP, the tight binding transition state analog carboxyarabinitol-1,5-bisphosphate, or inhibitory divalent metal ions within the active site. The catalytic properties of L(2) and L(10) M. burtonii Rubisco (MbR) were indistinguishable. At 25 degrees C they both shared a low specificity for CO(2) over O(2) (1.1 mol x mol(-1)) and RuBP carboxylation rates that were distinctively enhanced at low pH (approximately 4 s(-1) at pH 6, relative to 0.8 s(-1) at pH 8) with a temperature optimum of 55 degrees C. Like other archaeal Rubiscos, MbR also has a high O(2) affinity (K(m)(O(2)) = approximately 2.5 microM). The catalytic and structural similarities of MbR to other archaeal Rubiscos contrast with its closer sequence homology to bacterial L(2) Rubisco, complicating its classification within the Rubisco superfamily.
Figures



References
-
- Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Nat. Rev. Mol. Cell Biol. 5, 781–791 - PubMed
-
- Goloubinoff P., Christeller J. T., Gatenby A. A., Lorimer G. H. (1989) Nature 342, 884–889 - PubMed
-
- Andersson I., Backlund A. (2008) Plant Physiol. Biochem. 46, 275–291 - PubMed
-
- Brinker A., Pfeifer G., Kerner M. J., Naylor D. J., Hartl F. U., Hayer-Hartl M. (2001) Cell 107, 223–233 - PubMed
-
- Viitanen P. V., Lubben T. H., Reed J., Goloubinoff P., O'Keefe D. P., Lorimer G. H. (1990) Biochemistry 29, 5665–5671 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

