Autoaggregation response of Fusobacterium nucleatum
- PMID: 19837836
- PMCID: PMC2794088
- DOI: 10.1128/AEM.00916-09
Autoaggregation response of Fusobacterium nucleatum
Abstract
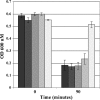
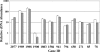
Fusobacterium nucleatum is a gram-negative oral bacterial species associated with periodontal disease progression. This species is perhaps best known for its ability to adhere to a vast array of other bacteria and eukaryotic cells. Numerous studies of F. nucleatum have examined various coaggregation partners and inhibitors, but it is largely unknown whether these interactions induce a particular genetic response. We tested coaggregation between F. nucleatum ATCC strain 25586 and various species of Streptococcus in the presence of a semidefined growth medium containing saliva. We found that this condition could support efficient coaggregation but, surprisingly, also stimulated a similar degree of autoaggregation. We further characterized the autoaggregation response, since few reports have examined this in F. nucleatum. After screening several common coaggregation inhibitors, we identified l-lysine as a competitive inhibitor of autoaggregation. We performed a microarray analysis of the planktonic versus autoaggregated cells and found nearly 100 genes that were affected after only about 60 min of aggregation. We tested a subset of these genes via real-time reverse transcription-PCR and confirmed the validity of the microarray results. Some of these genes were also found to be inducible in cell pellets created by centrifugation. Based upon these data, it appears that autoaggregation activates a genetic program that may be utilized for growth in a high cell density environment, such as the oral biofilm.
Figures





References
-
- Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1998. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol. Immunol. 13:51-54. - PubMed
-
- Bakken, V., B. T. Hogh, and H. B. Jensen. 1989. Utilization of amino acids and peptides by Fusobacterium nucleatum. Scand. J. Dent. Res. 97:43-53. - PubMed
-
- Bartold, P. M., N. J. Gully, P. S. Zilm, and A. H. Rogers. 1991. Identification of components in Fusobacterium nucleatum chemostat-culture supernatants that are potent inhibitors of human gingival fibroblast proliferation. J. Periodontal Res. 26:314-322. - PubMed
-
- Boiangiu, C. D., E. Jayamani, D. Brugel, G. Herrmann, J. Kim, L. Forzi, R. Hedderich, I. Vgenopoulou, A. J. Pierik, J. Steuber, and W. Buckel. 2005. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J. Mol. Microbiol. Biotechnol. 10:105-119. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

