Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature
- PMID: 19837982
- PMCID: PMC2830834
- DOI: 10.1074/mcp.M900152-MCP200
Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature
Abstract
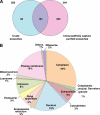
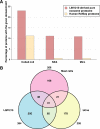
Exosomes are 40-100-nm-diameter nanovesicles of endocytic origin that are released from diverse cell types. To better understand the biological role of exosomes and to avoid confounding data arising from proteinaceous contaminants, it is important to work with highly purified material. Here, we describe an immunoaffinity capture method using the colon epithelial cell-specific A33 antibody to purify colorectal cancer cell (LIM1215)-derived exosomes. LC-MS/MS revealed 394 unique exosomal proteins of which 112 proteins (28%) contained signal peptides and a significant enrichment of proteins containing coiled coil, RAS, and MIRO domains. A comparative protein profiling analysis of LIM1215-, murine mast cell-, and human urine-derived exosomes revealed a subset of proteins common to all exosomes such as endosomal sorting complex required for transport (ESCRT) proteins, tetraspanins, signaling, trafficking, and cytoskeletal proteins. A conspicuous finding of this comparative analysis was the presence of host cell-specific (LIM1215 exosome) proteins such as A33, cadherin-17, carcinoembryonic antigen, epithelial cell surface antigen (EpCAM), proliferating cell nuclear antigen, epidermal growth factor receptor, mucin 13, misshapen-like kinase 1, keratin 18, mitogen-activated protein kinase 4, claudins (1, 3, and 7), centrosomal protein 55 kDa, and ephrin-B1 and -B2. Furthermore, we report the presence of the enzyme phospholipid scramblase implicated in transbilayer lipid distribution membrane remodeling. The LIM1215-specific exosomal proteins identified in this study may provide insights into colon cancer biology and potential diagnostic biomarkers.
Figures


References
-
- Simpson R. J., Jensen S. S., Lim J. W. ( 2008) Proteomic profiling of exosomes: current perspectives. Proteomics 8, 4083– 4099 - PubMed
-
- Taylor D. D., Akyol S., Gercel-Taylor C. ( 2006) Pregnancy-associated exosomes and their modulation of T cell signaling. J. Immunol 176, 1534– 1542 - PubMed
-
- Sabapatha A., Gercel-Taylor C., Taylor D. D. ( 2006) Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am. J. Reprod. Immunol 56, 345– 355 - PubMed
-
- Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., Tursz T., Amigorena S., Raposo G., Angevin E., Zitvogel L. ( 2002) Malignant effusions and immunogenic tumour-derived exosomes. Lancet 360, 295– 305 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

