Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription
- PMID: 19838172
- PMCID: PMC2912930
- DOI: 10.1038/ncb1984
Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription
Erratum in
- Nat Cell Biol. 2010 Nov;12(11):1122
Abstract
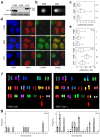
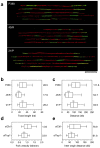
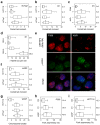
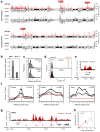
Topoisomerase I (Top1) is a key enzyme in functioning at the interface between DNA replication, transcription and mRNA maturation. Here, we show that Top1 suppresses genomic instability in mammalian cells by preventing a conflict between transcription and DNA replication. Using DNA combing and ChIP (chromatin immunoprecipitation)-on-chip, we found that Top1-deficient cells accumulate stalled replication forks and chromosome breaks in S phase, and that breaks occur preferentially at gene-rich regions of the genome. Notably, these phenotypes were suppressed by preventing the formation of RNA-DNA hybrids (R-loops) during transcription. Moreover, these defects could be mimicked by depletion of the splicing factor ASF/SF2 (alternative splicing factor/splicing factor 2), which interacts functionally with Top1. Taken together, these data indicate that Top1 prevents replication fork collapse by suppressing the formation of R-loops in an ASF/SF2-dependent manner. We propose that interference between replication and transcription represents a major source of spontaneous replication stress, which could drive genomic instability during the early stages of tumorigenesis.
Figures


References
-
- Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. - PubMed
-
- Ivessa AS, et al. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. - PubMed
-
- Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair. 2007;6:900–913. - PubMed
-
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
-
- Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

