The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA
- PMID: 19838203
- PMCID: PMC2847429
- DOI: 10.1038/nchembio.237
The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA
Abstract
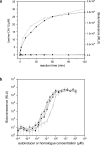
Vibrio cholerae, the bacterium that causes the disease cholera, controls virulence factor production and biofilm development in response to two extracellular quorum-sensing molecules, called autoinducers. The strongest autoinducer, called CAI-1 (for cholera autoinducer-1), was previously identified as (S)-3-hydroxytridecan-4-one. Biosynthesis of CAI-1 requires the enzyme CqsA. Here, we determine the CqsA reaction mechanism, identify the CqsA substrates as (S)-2-aminobutyrate and decanoyl coenzyme A, and demonstrate that the product of the reaction is 3-aminotridecan-4-one, dubbed amino-CAI-1. CqsA produces amino-CAI-1 by a pyridoxal phosphate-dependent acyl-CoA transferase reaction. Amino-CAI-1 is converted to CAI-1 in a subsequent step via a CqsA-independent mechanism. Consistent with this, we find cells release > or =100 times more CAI-1 than amino-CAI-1. Nonetheless, V. cholerae responds to amino-CAI-1 as well as CAI-1, whereas other CAI-1 variants do not elicit a quorum-sensing response. Thus, both CAI-1 and amino-CAI-1 have potential as lead molecules in the development of an anticholera treatment.
Figures


Comment in
-
Deciphering bacterial language.Nat Chem Biol. 2009 Dec;5(12):870-1. doi: 10.1038/nchembio.263. Nat Chem Biol. 2009. PMID: 19915530 No abstract available.
References
-
- Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. - PubMed
-
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. - PubMed
-
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–4. - PubMed
-
- Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–34. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- PubChem-Substance/85230822
- PubChem-Substance/85230823
- PubChem-Substance/85230824
- PubChem-Substance/85230825
- PubChem-Substance/85230826
- PubChem-Substance/85230827
- PubChem-Substance/85230828
- PubChem-Substance/85230829
- PubChem-Substance/85230830
- PubChem-Substance/85230831
- PubChem-Substance/85230832
- PubChem-Substance/85230833
- PubChem-Substance/85230834
- PubChem-Substance/85230835
- PubChem-Substance/85230836
- PubChem-Substance/85230837
- PubChem-Substance/85230838
- PubChem-Substance/85230839
- PubChem-Substance/85230840
- PubChem-Substance/85230841
- PubChem-Substance/85230842
- PubChem-Substance/85230843
- PubChem-Substance/85230844
- PubChem-Substance/85230845
- PubChem-Substance/85230846
- PubChem-Substance/85230847
- PubChem-Substance/85230848
- PubChem-Substance/85230849
- PubChem-Substance/85230850
- PubChem-Substance/85230851
- PubChem-Substance/85230852
- PubChem-Substance/85230853
- PubChem-Substance/85230854
- PubChem-Substance/85230855
- PubChem-Substance/85230856
- PubChem-Substance/85230857
- PubChem-Substance/85230858
- PubChem-Substance/85230859
- PubChem-Substance/85230860
- PubChem-Substance/85230861
- PubChem-Substance/85230862
- PubChem-Substance/85230863
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

