Cohesin SMC1beta protects telomeres in meiocytes
- PMID: 19841137
- PMCID: PMC2768837
- DOI: 10.1083/jcb.200808016
Cohesin SMC1beta protects telomeres in meiocytes
Abstract
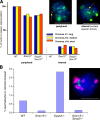
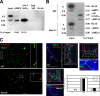
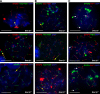
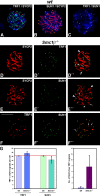
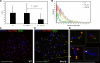
Meiosis-specific mammalian cohesin SMC1beta is required for complete sister chromatid cohesion and proper axes/loop structure of axial elements (AEs) and synaptonemal complexes (SCs). During prophase I, telomeres attach to the nuclear envelope (NE), but in Smc1beta(-/-) meiocytes, one fifth of their telomeres fail to attach. This study reveals that SMC1beta serves a specific role at telomeres, which is independent of its role in determining AE/SC length and loop extension. SMC1beta is necessary to prevent telomere shortening, and SMC3, present in all known cohesin complexes, properly localizes to telomeres only if SMC1beta is present. Very prominently, telomeres in Smc1beta(-/-) spermatocytes and oocytes loose their structural integrity and suffer a range of abnormalities. These include disconnection from SCs and formation of large telomeric protein-DNA extensions, extended telomere bridges between SCs, ring-like chromosomes, intrachromosomal telomeric repeats, and a reduction of SUN1 foci in the NE. We suggest that a telomere structure protected from DNA rearrangements depends on SMC1beta.
Figures




References
-
- Achi M.V., Ravindranath N., Dym M. 2000. Telomere length in male germ cells is inversely correlated with telomerase activity.Biol. Reprod. 63:591–598 doi:10.1095/biolreprod63.2.591 - DOI - PubMed
-
- Akhmedov A.T., Gross B., Jessberger R. 1999. Mammalian SMC3 C-terminal and coiled-coil protein domains specifically bind palindromic DNA, do not block DNA ends, and prevent DNA bending.J. Biol. Chem. 274:38216–38224 doi:10.1074/jbc.274.53.38216 - DOI - PubMed
-
- Bannister L.A., Schimenti J.C. 2004. Homologous recombinational repair proteins in mouse meiosis.Cytogenet. Genome Res. 107:191–200 doi:10.1159/000080597 - DOI - PubMed
-
- Bastos H., Lassalle B., Chicheportiche A., Riou L., Testart J., Allemand I., Fouchet P. 2005. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis.Cytometry A. 65:40–49 - PubMed
-
- Blackburn E.H. 2005. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions.FEBS Lett. 579:859–862 doi:10.1016/j.febslet.2004.11.036 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

